| MC JC |
AE015451_phaM1 |
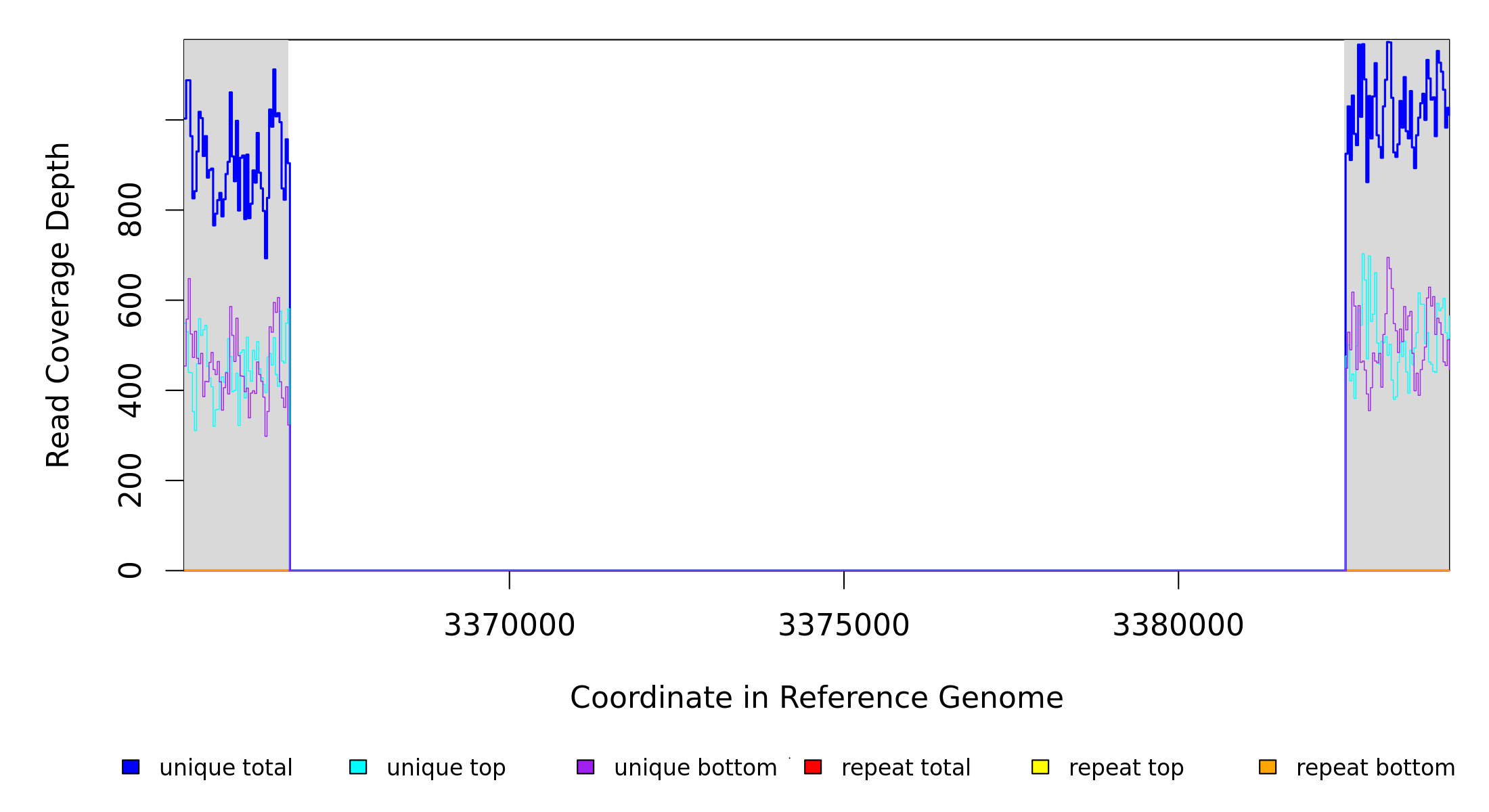
3,366,695 |
Δ15,777 bp |
|
[PP_2964 CDS (putative transposase)]–[PP_2984 CDS (conserved protein of unknown function)] |
20 genes [PP_2964 CDS (putative transposase)], tnpC CDS (putative tnpA repressor TnpC), PP_2968 CDS (putative nickel_cobalt transporter_ high‑affinity), csoR CDS (putative repressor of copper utilisation proteins), PP_2970 CDS (conserved exported protein of unknown function), PP_2971 CDS (transposase), PP_2972 CDS (conserved protein of unknown function), dgkA‑II CDS (diacylglycerol kinase), PP_2974 CDS (putative sulfatase), PP_2975 CDS (Insertion element ISR1 uncharacterized 10 kDa protein A3), PP_2976 CDS (putative transposase), PP_mr38 ncRNA (ALIL regulatory region), tnpB CDS (Tn4652_ transposase subunit B), PP_2978 CDS (conserved protein of unknown function), PP_2979 CDS (conserved protein of unknown function), PP_2980 CDS (conserved protein of unknown function), tnpS CDS (putative cointegrate resolution protein S), tnpT‑I CDS (putative cointegrate resolution protein T Tn4652), PP_2983 CDS (conserved protein of unknown function), [PP_2984 CDS (conserved protein of unknown function)] |