| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
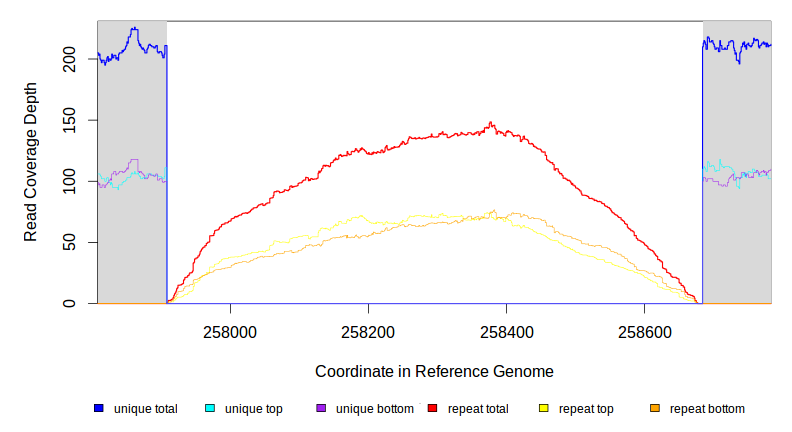
| MC JC | NC_000913 | 257,908 | Δ776 bp | [crl] | [crl] | |
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_000913 | 257908–258676 | 258683 | 8–776 | 211 [0] | [0] 210 | [crl] | [crl] |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_000913 | = 257907 | 0 (0.000) | 198 (0.950) | 100/432 | 0.4 | 100% | intergenic (+8/‑769) | crl/crl | pseudogene, sigma factor‑binding protein, RNA polymerase holoenzyme formation stimulator;regulator; Surface structures; transcriptional regulator of cryptic csgA gene for curli surface fibers/pseudogene, sigma factor‑binding protein, RNA polymerase holoenzyme formation stimulator;regulator; Surface structures; transcriptional regulator of cryptic csgA gene for curli surface fibers |
| ? | NC_000913 | 258684 = | 0 (0.000) | pseudogene (9/331 nt) | crl | pseudogene, sigma factor‑binding protein, RNA polymerase holoenzyme formation stimulator;regulator; Surface structures; transcriptional regulator of cryptic csgA gene for curli surface fibers | |||||
