| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
| MC JC | NC_000913 | 700,785 | Δ47 bp | coding (764‑810/1221 nt) | nagC ← | N‑acetylglucosamine‑inducible nag divergent operon transcriptional repressor |
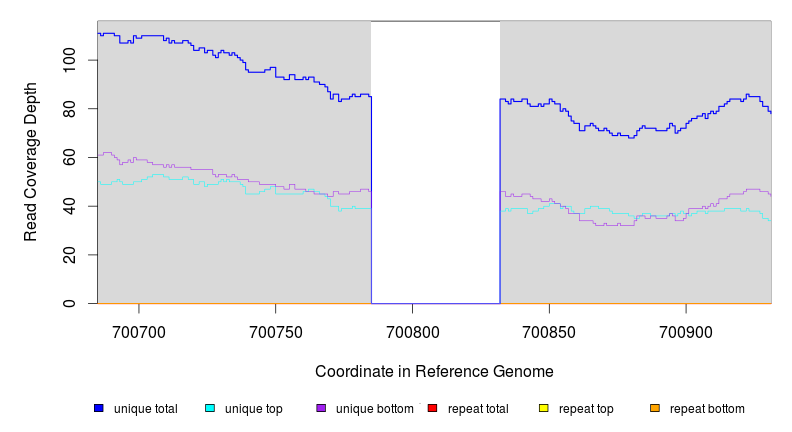
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_000913 | 700785 | 700831 | 47 | 85 [0] | [0] 84 | nagC | N‑acetylglucosamine‑inducible nag divergent operon transcriptional repressor |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_000913 | = 700784 | 0 (0.000) | 80 (0.920) | 66/278 | 0.5 | 100% | coding (811/1221 nt) | nagC | N‑acetylglucosamine‑inducible nag divergent operon transcriptional repressor |
| ? | NC_000913 | 700832 = | 0 (0.000) | coding (763/1221 nt) | nagC | N‑acetylglucosamine‑inducible nag divergent operon transcriptional repressor | |||||
