Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate | Predicted Mutations | Mean Coverage | Total Reads | Percent Mapped | Mapped Reads | Average Read Length |
|---|---|---|---|---|---|---|
| A2 F57 I1 R1 | 57 | 81.4 | 3444058 | 98.4% | 3388953 | 146.5 |
Breseq alignment
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
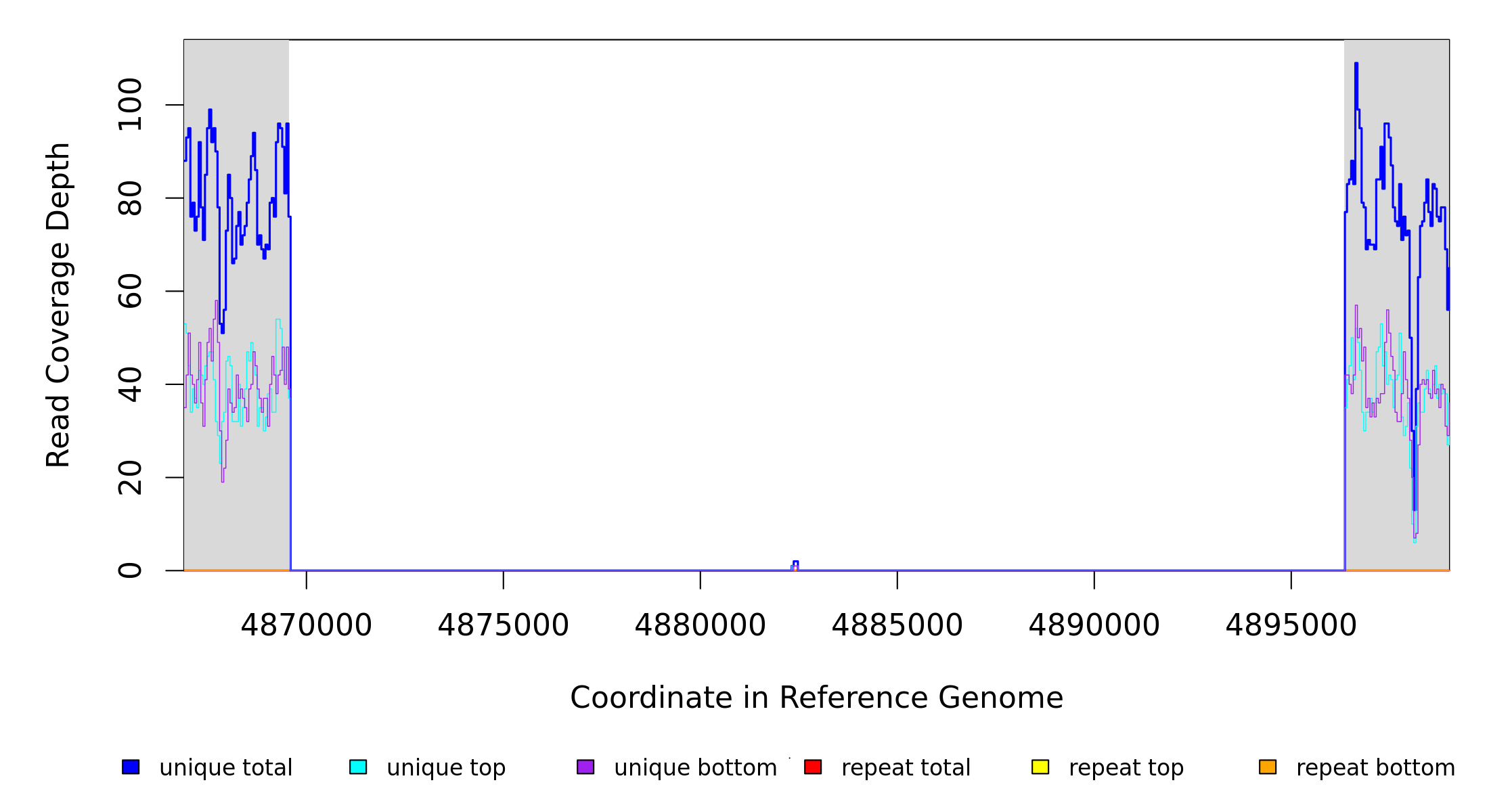
| MC JC | NC_002947_CJ‑RC | 4,869,553 | Δ26,791 bp | [xdhA]–PP_4302 | 24 genes [xdhA], xdhB, xdhC, guaD, aqpZ, PP_4283, PP_4284, pucM, puuE, pucL, allA, PP_4289, uacT, PP_4291, PP_4293, yedI, PP_4295, PP_4296, gcl, hyi, glxR, ttuD, pyk, PP_4302 |
|
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_002947_CJ‑RC | 4869553 | 4896343 | 26791 | 76 [0] | [0] 76 | [xdhA]–PP_4302 | [xdhA],xdhB,xdhC,guaD,aqpZ,PP_4283,PP_4284,pucM,puuE,pucL,allA,PP_4289,uacT,PP_4291,PP_4293,yedI,PP_4295,PP_4296,gcl,hyi,glxR,ttuD,pyk,PP_4302 |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_002947_CJ‑RC | = 4869552 | 0 (0.000) | 75 (0.950) | 63/288 | 0.2 | 100% | coding (1046/1455 nt) | xdhA | xanthine dehydrogenase subunit XdhA |
| ? | NC_002947_CJ‑RC | 4896344 = | 0 (0.000) | intergenic (+33/+18) | PP_4302/PP_4303 | urea transporter/hypothetical protein | |||||