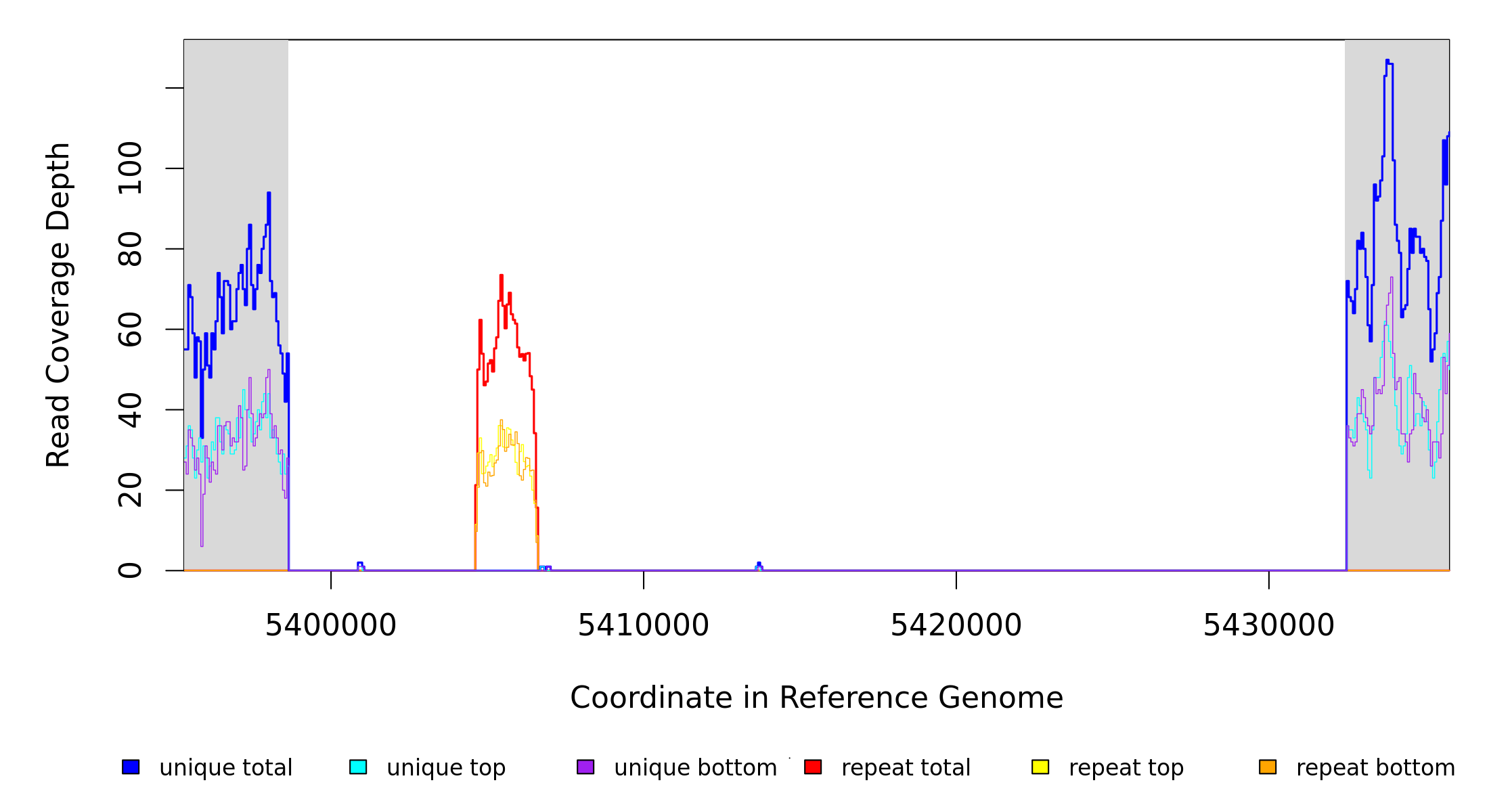
Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate | Predicted Mutations | Mean Coverage | Total Reads | Percent Mapped | Mapped Reads | Average Read Length |
|---|---|---|---|---|---|---|
| A5 F26 I1 R1 | 57 | 84.6 | 3443374 | 98.5% | 3391723 | 147.1 |
Breseq alignment
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
| MC JC | NC_002947_CJ‑RC | 5,398,630 | Δ33,798 bp | [hsdM]–[PP_4768] | 28 genes [hsdM], hsdS, PP_5711, PP_4743, PP_4745, PP_4746, PP_4748, PP_4749, PP_4750, PP_4751, PP_4752, stcD, PP_4754, PP_4755, gabP‑V, gudD, PP_4758, PP_4759, PP_4760, PP_4761, tesB, PP_4763, PP_5712, PP_4764, PP_4765, rhlE‑I, yedA, [PP_4768] |
|
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_002947_CJ‑RC | 5398630 | 5432427 | 33798 | 61 [0] | [0] 62 | [hsdM]–[PP_4768] | [hsdM],hsdS,PP_5711,PP_4743,PP_4745,PP_4746,PP_4748,PP_4749,PP_4750,PP_4751,PP_4752,stcD,PP_4754,PP_4755,gabP‑V,gudD,PP_4758,PP_4759,PP_4760,PP_4761,tesB,PP_4763,PP_5712,PP_4764,PP_4765,rhlE‑I,yedA,[PP_4768] |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_002947_CJ‑RC | = 5398629 | 0 (0.000) | 61 (0.770) | 52/288 | 0.6 | 100% | coding (40/1218 nt) | hsdM | type I restriction modification system methylase |
| ? | NC_002947_CJ‑RC | 5432428 = | 0 (0.000) | coding (217/612 nt) | PP_4768 | exonuclease | |||||