| MC JC |
NC_002947_CJ‑RC |
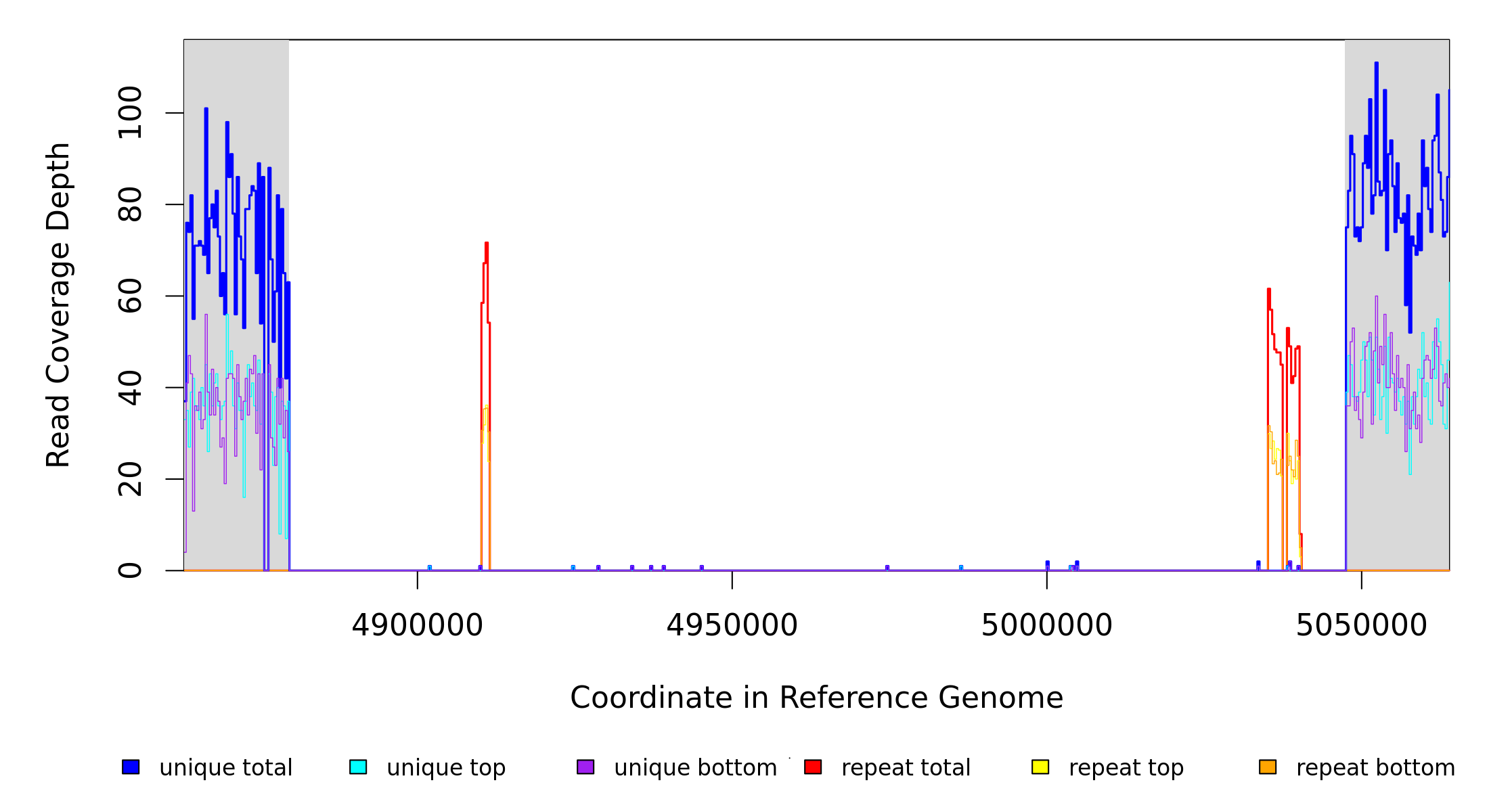
4,879,568 |
Δ167,714 bp |
|
[puuE]–[PP_4449] |
164 genes [puuE], pucL, allA, PP_4289, uacT, PP_4291, PP_4293, yedI, PP_4295, PP_4296, gcl, hyi, glxR, ttuD, pyk, PP_4302, PP_4303, PP_4304, sbp‑I, PP_4306, PP_4307, PP_4308, PP_4309, PP_4310, PP_4311, PP_4312, PP_4313, PP_4314, PP_4315, PP_4316, PP_4317, PP_4318, PP_4319, cycH, ccmH, ccmG, ccmF, ccmE, ccmD, ccmC, ccmB, ccmA, PP_4328, PP_4329, PP_4330, PP_4331, PP_4332, PP_4333, PP_4334, PP_4335, PP_4336, cheBA, cheA, cheZ, cheY, fliA, PP_4342, flhF, flhA, PP_4345, ddlA, PP_4348, PP_4349, PP_4350, PP_5673, flhB, fliR, fliQ, fliP, fliO, fliN, fliM, fliL, fliK, PP_4362, PP_4363, PP_4364, fliJ, fliI, fliH, fliG, fliF, fliE, atoC, fleS, fleQ, fliT, fliS, fliD, PP_4377, fliC, PP_4379, flgL, flgK, flgJ, flgI, flgH, flgG, flgF, PP_4387, flgE, flgD, flgC, flgB, cheR, PP_4393, flgA, flgM, PP_4396, ycgR, PP_4398, PP_4399, bkdR, bkdAA, bkdAB, bkdB, lpdV, PP_4405, PP_4406, PP_4407, PP_4409, PP_4410, PP_5674, PP_4411, PP_4412, PP_4413, PP_4415, PP_5675, PP_5676, PP_5677, PP_5678, PP_4418, insN, PP_4420, PP_5679, PP_4421, gabD‑II, PP_4423, PP_4424, PP_4425, PP_4426, PP_4427, PP_4428, PP_4429, PP_4430, ocd, PP_4432, PP_4433, dadA‑I, PP_4435, PP_4437, PP_4438, PP_4439, PP_4441, PP_4442, PP_4443, PP_4444, PP_4445, PP_4446, PP_5680, PP_4447, PP_4448, PP_5681, [PP_4449] |