Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A0 F0 I1 R1
|
245 |
579.6 |
28867798 |
97.7% |
28203838 |
96.0 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
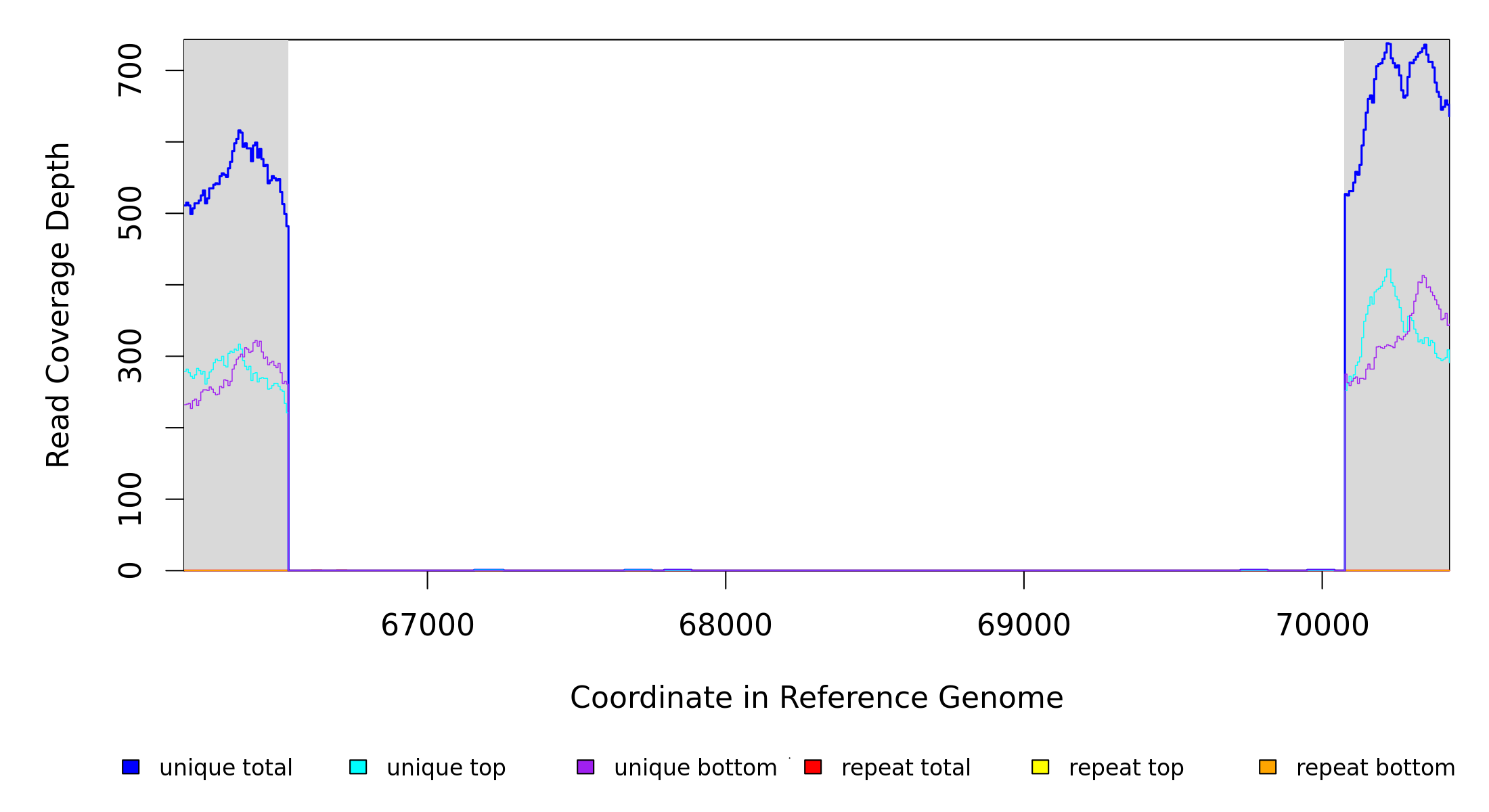
NC_000913 |
66,533 |
3540 bp→CCCGAGACCCGCACGCGTCGACCTGCA |
|
[araD]–araB |
[araD], araA, araB |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
66533 |
70072 |
3540 |
478 [0] |
[0] 527 |
[araD]–araB |
[araD],araA,araB |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 66532 | 0 (0.000) | 337 (0.820)
+CCCGAGACCCGCACGCGTCGACCTGCA |
112/136 |
1.3 |
100% |
coding (19/696 nt) |
araD |
L‑ribulose‑5‑phosphate 4‑epimerase |
| ? | NC_000913 |
70073 = |
0 (0.000) | intergenic (‑25/‑314) |
araB/araC |
L‑ribulokinase/transcriptional regulator for ara operon |
GATK/CNVnator alignment
N/A