Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate | Predicted Mutations | Mean Coverage | Total Reads | Percent Mapped | Mapped Reads | Average Read Length |
|---|---|---|---|---|---|---|
| A3 F55 I1 R1 | 61 | 67.4 | 2858014 | 98.3% | 2809427 | 146.5 |
Breseq alignment
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
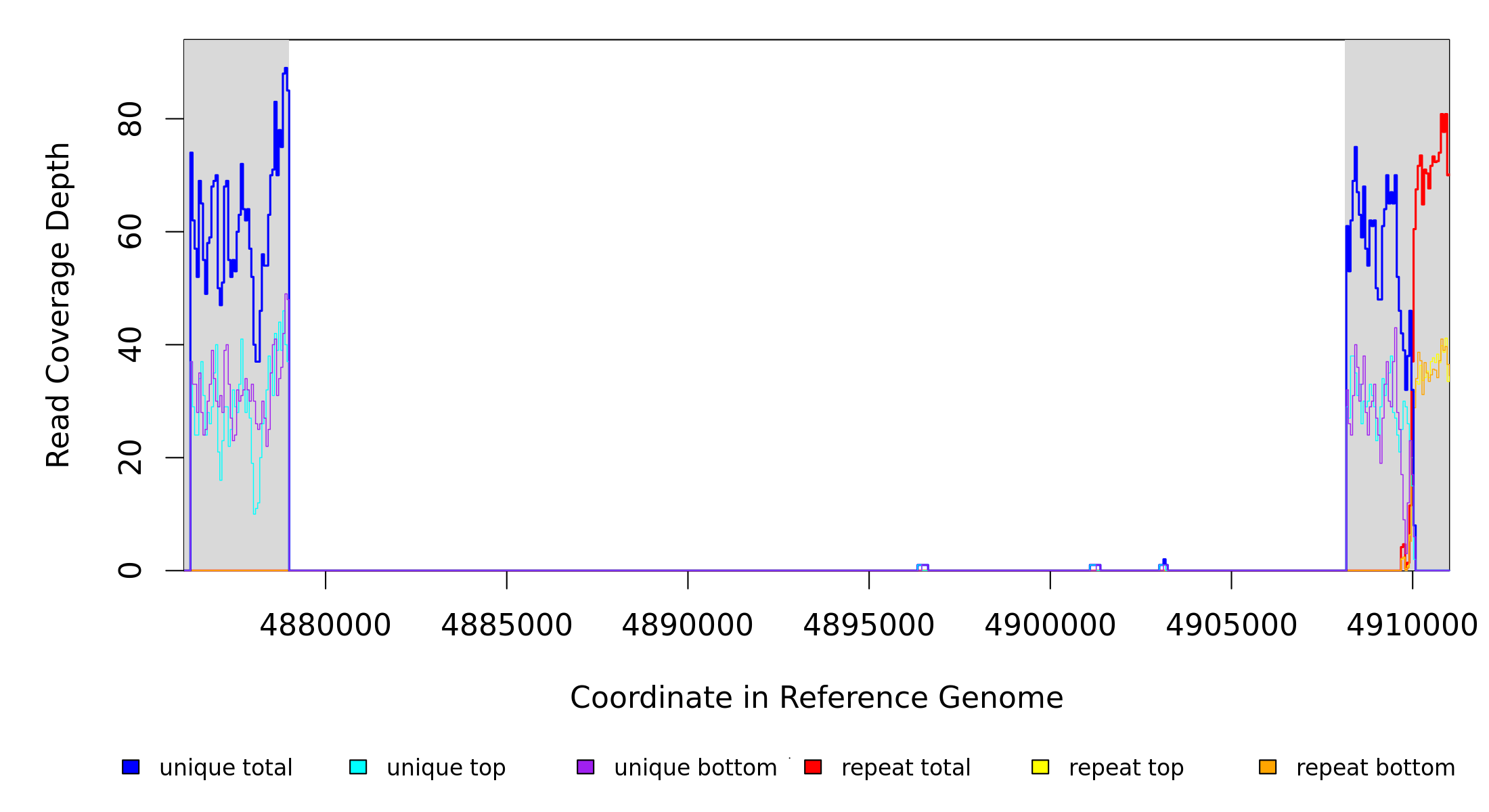
| MC JC | NC_002947_CJ‑RC | 4,878,995 | Δ29,127 bp | puuE–[PP_4315] | 29 genes puuE, pucL, allA, PP_4289, uacT, PP_4291, PP_4293, yedI, PP_4295, PP_4296, gcl, hyi, glxR, ttuD, pyk, PP_4302, PP_4303, PP_4304, sbp‑I, PP_4306, PP_4307, PP_4308, PP_4309, PP_4310, PP_4311, PP_4312, PP_4313, PP_4314, [PP_4315] |
|
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_002947_CJ‑RC | 4878995 | 4908121 | 29127 | 64 [0] | [0] 63 | puuE–[PP_4315] | puuE,pucL,allA,PP_4289,uacT,PP_4291,PP_4293,yedI,PP_4295,PP_4296,gcl,hyi,glxR,ttuD,pyk,PP_4302,PP_4303,PP_4304,sbp‑I,PP_4306,PP_4307,PP_4308,PP_4309,PP_4310,PP_4311,PP_4312,PP_4313,PP_4314,[PP_4315] |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_002947_CJ‑RC | = 4878994 | 0 (0.000) | 62 (0.940) | 53/288 | 0.3 | 100% | intergenic (‑339/‑61) | pucM/puuE | 5‑hydroxyisourate hydrolase/allantoinase |
| ? | NC_002947_CJ‑RC | 4908122 = | 0 (0.000) | coding (379/789 nt) | PP_4315 | epimerase | |||||