Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A3 F244 I1 R1
|
11 |
176.3 |
4507478 |
97.4% |
4390283 |
183.3 |
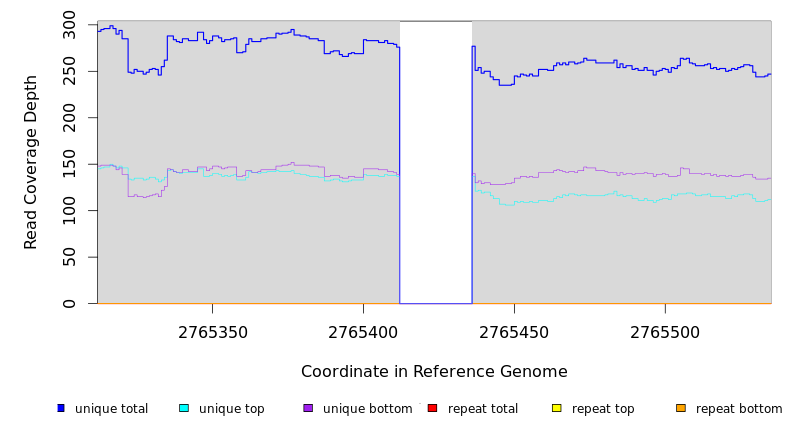
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
2,765,440 |
(TATGGCAC)6→3 |
intergenic (‑287/+50) |
yfjL ← / ← yfjM |
CP4‑57 putative defective prophage, DUF4297/DUF1837 polymorphic toxin family protein/CP4‑57 prophage; uncharacterized protein |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
2765412 |
2765435 |
24 |
276 [0] |
[0] 277 |
yfjL/yfjM |
CP4‑57 putative defective prophage, DUF4297/DUF1837 polymorphic toxin family protein/CP4‑57 prophage; uncharacterized protein |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
2765436 = | 0 (0.000) | 210 (1.450) |
103/308 |
0.0 |
100% |
intergenic (‑283/+77) |
yfjL/yfjM |
CP4‑57 putative defective prophage, DUF4297/DUF1837 polymorphic toxin family protein/CP4‑57 prophage; uncharacterized protein |
| ? | NC_000913 |
= 2765411 |
0 (0.000) | intergenic (‑258/+102) |
yfjL/yfjM |
CP4‑57 putative defective prophage, DUF4297/DUF1837 polymorphic toxin family protein/CP4‑57 prophage; uncharacterized protein |
GATK/CNVnator alignment
N/A