Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A2 F1 I2 R1
|
764 |
42.0 |
2018750 |
95.5% |
1927906 |
109.2 |
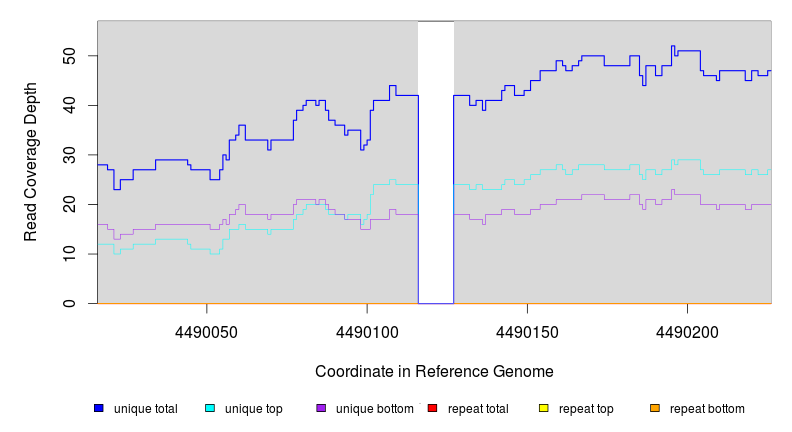
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,490,116 |
Δ11 bp |
intergenic (‑53/+15) |
yjgR ← / ← idnR |
DUF853 family protein with NTPase fold/transcriptional repressor, 5‑gluconate‑binding |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4490116 |
4490126 |
11 |
42 [0] |
[0] 42 |
yjgR/idnR |
DUF853 family protein with NTPase fold/transcriptional repressor, 5‑gluconate‑binding |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4490115 | 0 (0.000) | 42 (0.950) |
35/216 |
0.2 |
100% |
intergenic (‑52/+26) |
yjgR/idnR |
DUF853 family protein with NTPase fold/transcriptional repressor, 5‑gluconate‑binding |
| ? | NC_000913 |
4490127 = |
0 (0.000) | intergenic (‑64/+14) |
yjgR/idnR |
DUF853 family protein with NTPase fold/transcriptional repressor, 5‑gluconate‑binding |
GATK/CNVnator alignment
N/A