Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate | Predicted Mutations | Mean Coverage | Total Reads | Percent Mapped | Mapped Reads | Average Read Length |
|---|---|---|---|---|---|---|
| A11 F60 I1 R1 | 17 | 124.2 | 4215036 | 99.0% | 4172885 | 138.1 |
Breseq alignment
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
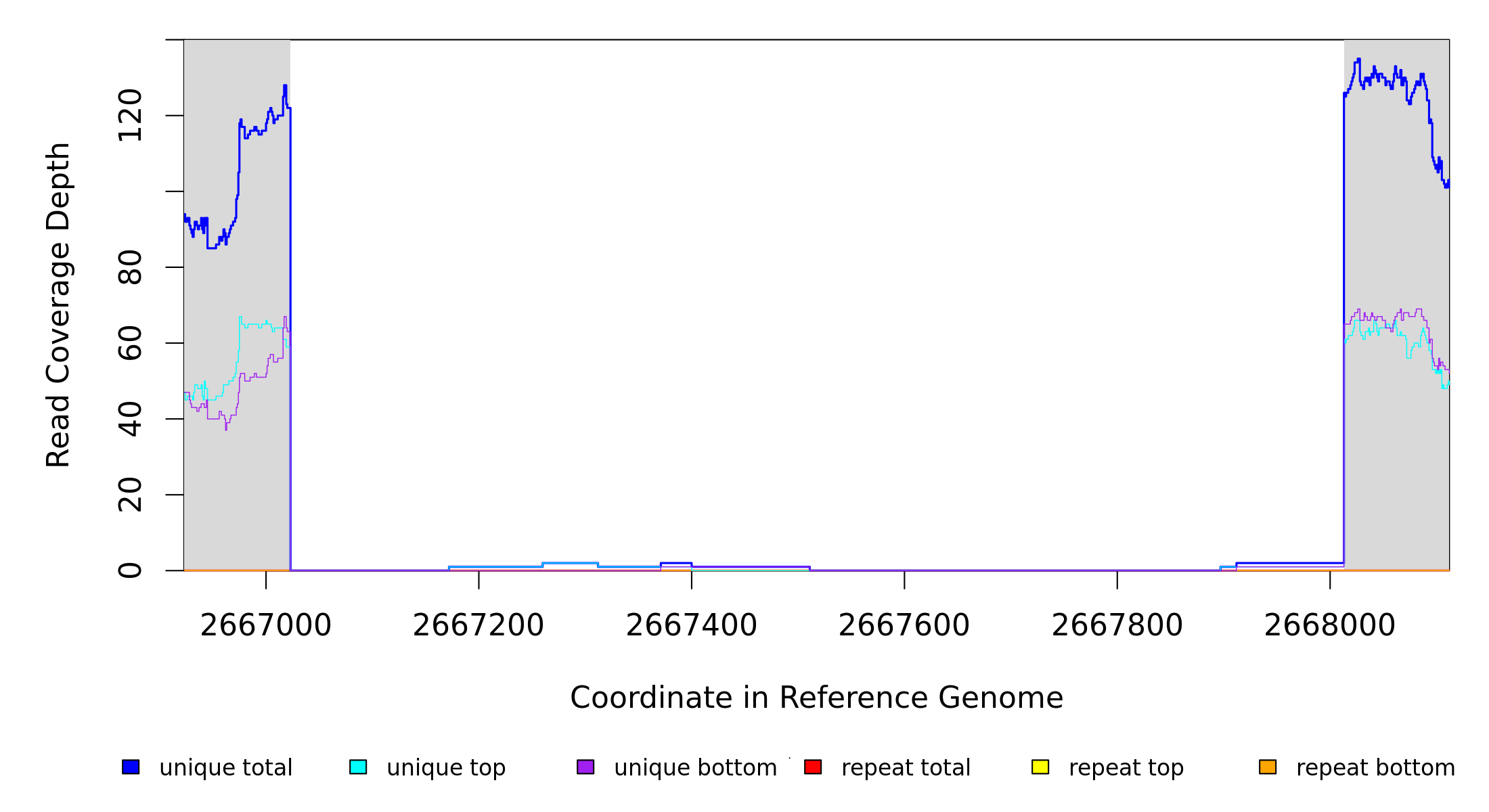
| MC JC | NC_000913 | 2,667,023 | Δ990 bp | [hcaT]–[hcaR] | [hcaT], [iroK], [hcaR] | |
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_000913 | 2667023 | 2668012 | 990 | 122 [0] | [2] 126 | [hcaT]–[hcaR] | [hcaT],[iroK],[hcaR] |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_000913 | = 2667022 | 0 (0.000) | 121 (0.980) | 65/274 | 0.4 | 99.2% | coding (825/1140 nt) | hcaT | putative 3‑phenylpropionate transporter |
| ? | NC_000913 | 2668013 = | 2 (0.020) | coding (59/66 nt) coding (884/891 nt) |
iroK hcaR |
protein IroK DNA‑binding transcriptional dual regulator HcaR |
|||||