Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A11 F64 I0 R1
|
613 |
127.6 |
4037778 |
97.7% |
3944909 |
149.9 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
freq |
annotation |
gene |
description |
| MC JC |
NC_000913 |
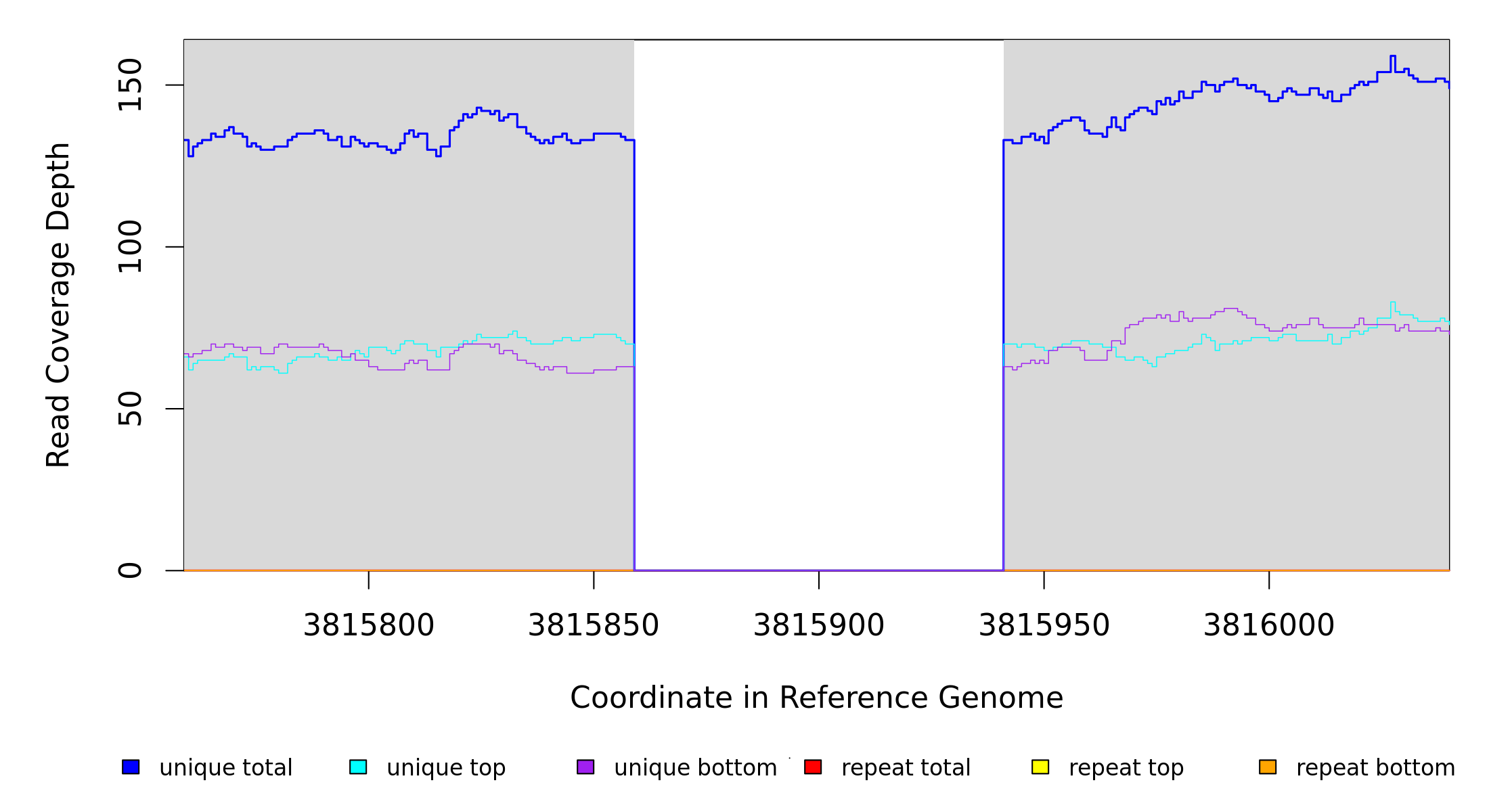
3,815,859 |
Δ82 bp |
100% |
|
[rph]–[rph] |
[rph], [rph] |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
3815859 |
3815940 |
82 |
133 [0] |
[0] 133 |
[rph]–[rph] |
[rph],[rph] |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 3815858 | 0 (0.000) | 122 (1.120) |
86/258 |
NT |
100% |
pseudogene (24/48 nt) |
rph |
ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH |
| ? | NC_000913 |
3815941 = |
0 (0.000) | pseudogene (609/669 nt) |
rph |
ribonuclease PH (defective);enzyme; Degradation of RNA; RNase PH |
GATK/CNVnator alignment
N/A