Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A0 F0 I1 R1
|
146 |
27.9 |
2327082 |
89.7% |
2087392 |
107.8 |
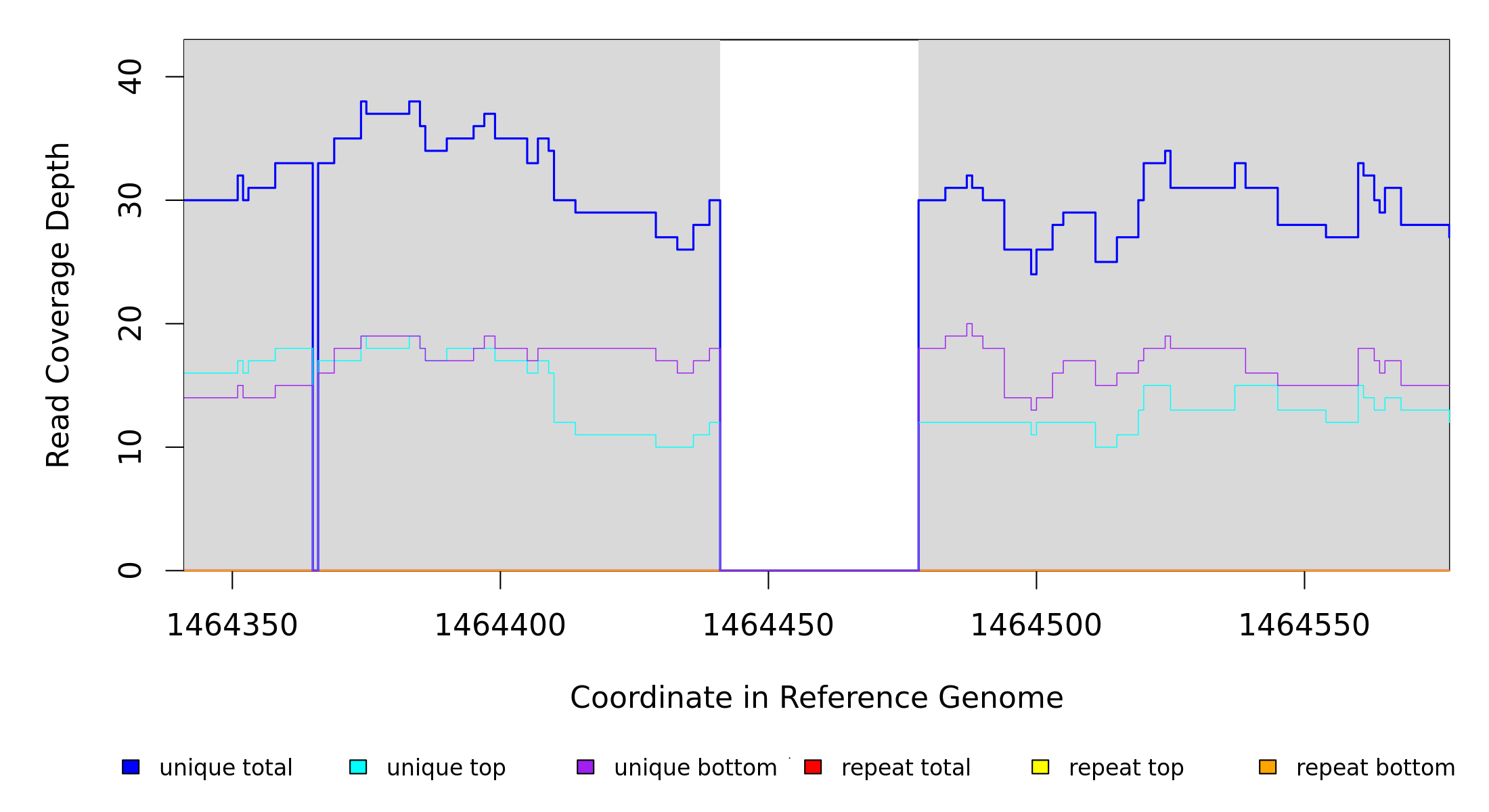
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
AM260479 |
1,464,441 |
Δ37 bp |
coding (125‑161/552 nt) |
h16_A1354 → |
conserved hypothetical protein |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
AM260479 |
1464441 |
1464477 |
37 |
30 [0] |
[0] 30 |
h16_A1354 |
conserved hypothetical protein |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
AM260479 |
= 1464440 | 0 (0.000) | 30 (1.090) |
21/214 |
0.3 |
100% |
coding (124/552 nt) |
h16_A1354 |
conserved hypothetical protein |
| ? | AM260479 |
1464478 = |
0 (0.000) | coding (162/552 nt) |
h16_A1354 |
conserved hypothetical protein |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
CGCCGTGACAGGAGATGCCATGCACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGCAGTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAACTGCTGGTGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGGCATTGGGTGAGCCGCCGAATATGTGGCGAATGGGCCCGCGCCCGGTCGACTGGCCTGATAA > AM260479/1464298‑1464578
|
CGCCGTGACAGGAGATGCCATGCACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCAC < GWNJ‑0478:712:GW2002102894th:2:2215:6822:72158/150‑1 (MQ=60)
GCCATGCACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGG < GWNJ‑0478:712:GW2002102894th:2:1204:19914:53368/150‑1 (MQ=60)
CCATGCACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGA < GWNJ‑0478:712:GW2002102894th:2:1111:7635:76758/150‑1 (MQ=60)
ATGCCCGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATG < GWNJ‑0478:712:GW2002102894th:2:2205:1767:32687/150‑1 (MQ=60)
ACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACC < GWNJ‑0478:712:GW2002102894th:2:1103:18779:25999/150‑1 (MQ=60)
ACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACC < GWNJ‑0478:712:GW2002102894th:2:2205:17794:51487/150‑1 (MQ=60)
GTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGC < GWNJ‑0478:712:GW2002102894th:2:2201:18951:8958/149‑1 (MQ=60)
CATCAGCTGGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGgcattgggt > GWNJ‑0478:712:GW2002102894th:2:2201:15324:19331/1‑141 (MQ=60)
gagatgtgtataagagacaggtccagcGTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGgcattgggt < GWNJ‑0478:712:GW2002102894th:2:2209:1567:15644/123‑10 (MQ=60)
GGTGGGCGAACCGTCCAGC‑GTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGGCGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGgcattgggtctgtctct > GWNJ‑0478:712:GW2002102894th:2:1210:5378:93042/1‑133 (MQ=60)
CTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAGCCGCTGG‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑CGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGGCATTGGGTGAGCCGCCGAATATctgtctcttatacacatctccgagcccacgagacactcg > GWNJ‑0478:712:GW2002102894th:2:2111:19726:94460/1‑111 (MQ=57)
|
CGCCGTGACAGGAGATGCCATGCACGCCTGTGACCTGTGTCATCAGCTGGTGGGCGAACCGTCCAGCAGTGCCGCCACACGAGCACCTGGCTGCCTCTGGCATTGCGGCCATGAGCAACCGGTGCAAGGTCGACAACTGCTGGTGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGCTGCACGGGCTGCGGCGCCTGGATGTACCAGAGCACGGCATTGGGTGAGCCGCCGAATATGTGGCGAATGGGCCCGCGCCCGGTCGACTGGCCTGATAA > AM260479/1464298‑1464578
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 30 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG < 40 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |