Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A0 F0 I1 R1
|
61 |
39.0 |
4165548 |
97.4% |
4057243 |
49.6 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
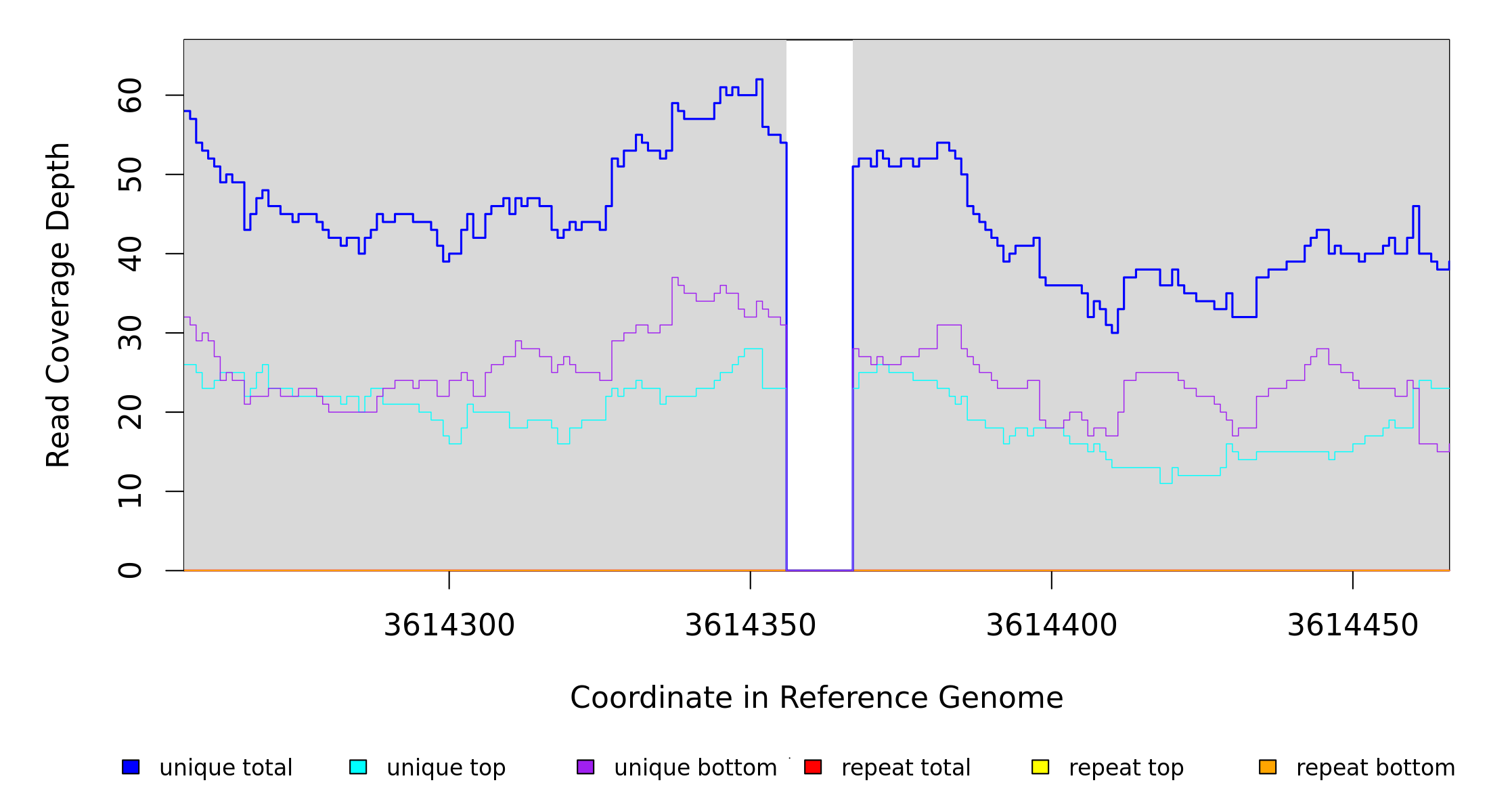
NC_017635 |
3,614,356 |
Δ11 bp |
coding (1926‑1936/1941 nt) |
csrD ← |
RNase E specificity factor CsrD |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_017635 |
3614356 |
3614366 |
11 |
54 [0] |
[0] 51 |
csrD |
RNase E specificity factor CsrD |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_017635 |
= 3614355 | 0 (0.000) | 51 (1.260) |
37/98 |
0.1 |
100% |
coding (1937/1941 nt) |
csrD |
RNase E specificity factor CsrD |
| ? | NC_017635 |
3614367 = |
0 (0.000) | coding (1925/1941 nt) |
csrD |
RNase E specificity factor CsrD |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
GACGCAGCGCGCGTTATTCTACGTGAAAACAGATTAAACGGCAGGTTAAACCGAGTATCTTTGTGAATATTTTTTCACGTTAGTATCAAGTGGCTGTGA > NC_017635/3614306‑3614404
|
GACGCAGCGCGCGTTATTCTACGTGAAAACAGATTAAACGGCAGGTTAAAT < M01420:85:000000000‑AC936:1:1110:26991:16304/51‑1 (MQ=60)
TTATTCTACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTT > M01420:85:000000000‑AC936:1:2114:21998:4557/1‑51 (MQ=41)
TATTCTACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTC > M01420:85:000000000‑AC936:1:2109:19146:27251/1‑51 (MQ=38)
TATTCTACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTC > M01420:85:000000000‑AC936:1:2103:7593:13649/1‑51 (MQ=38)
TCTACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACG > M01420:85:000000000‑AC936:1:2111:18069:25980/1‑51 (MQ=33)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTT > M01420:85:000000000‑AC936:1:1101:23904:16927/1‑50 (MQ=31)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTT > M01420:85:000000000‑AC936:1:1110:7383:17969/1‑50 (MQ=31)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTT > M01420:85:000000000‑AC936:1:1101:18585:5655/1‑50 (MQ=31)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGT < M01420:85:000000000‑AC936:1:1102:20349:27727/49‑1 (MQ=28)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACG < M01420:85:000000000‑AC936:1:2110:13612:14201/48‑1 (MQ=26)
ACGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACG < M01420:85:000000000‑AC936:1:2110:9968:14630/48‑1 (MQ=26)
CGTGAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTT > M01420:85:000000000‑AC936:1:2105:9753:26130/1‑49 (MQ=28)
GAAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTA < M01420:85:000000000‑AC936:1:2106:9910:4234/50‑1 (MQ=31)
AAAACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATC > M01420:85:000000000‑AC936:1:2113:11418:12688/1‑51 (MQ=33)
ACAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAG < M01420:85:000000000‑AC936:1:2102:14257:19810/51‑1 (MQ=33)
CAGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGT > M01420:85:000000000‑AC936:1:2114:18131:24492/1‑51 (MQ=33)
AGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGTG < M01420:85:000000000‑AC936:1:2104:25093:18056/51‑1 (MQ=33)
AGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGTG < M01420:85:000000000‑AC936:1:2105:19499:5316/51‑1 (MQ=33)
AGATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGTG < M01420:85:000000000‑AC936:1:2111:9293:15431/51‑1 (MQ=33)
GATTAAACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGTGG < M01420:85:000000000‑AC936:1:1110:12160:5356/51‑1 (MQ=33)
ACGGCAGGTTAAA‑‑‑‑‑‑‑‑‑‑‑TGTGAATATTTTTTCACGTTAGTATCAAGTGGCTGTGA < M01420:85:000000000‑AC936:1:1102:18125:16444/51‑1 (MQ=44)
|
GACGCAGCGCGCGTTATTCTACGTGAAAACAGATTAAACGGCAGGTTAAACCGAGTATCTTTGTGAATATTTTTTCACGTTAGTATCAAGTGGCTGTGA > NC_017635/3614306‑3614404
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 33 ≤ ATCG/ATCG < 37 ≤ ATCG/ATCG < 39 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |