Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A1 F2 I216 R1
|
8 |
28.7 |
1753078 |
96.2% |
1686461 |
85.5 |
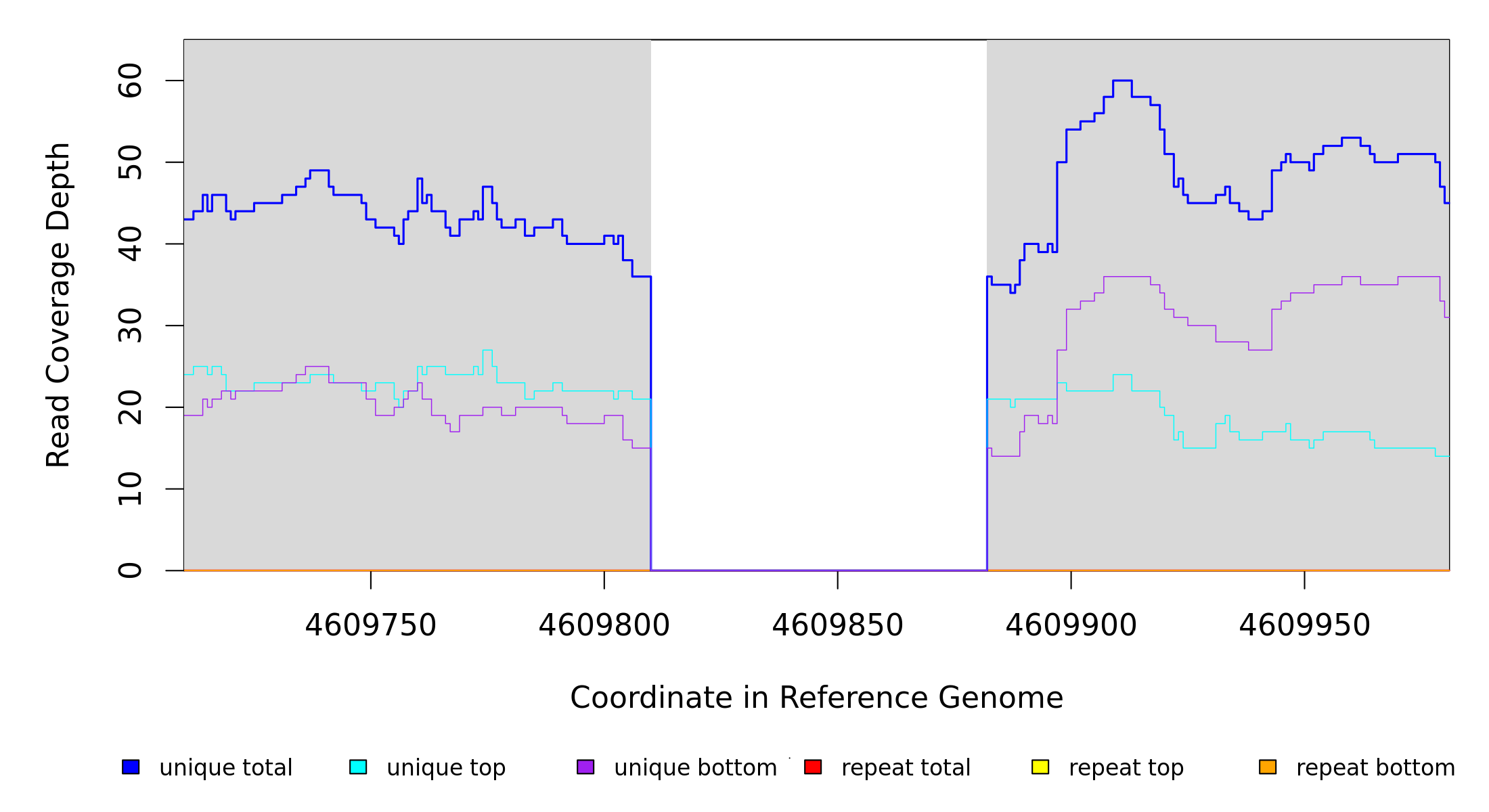
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NZ_CP009273 |
4,609,810 |
Δ72 bp |
coding (391‑462/1224 nt) |
deoB → |
phosphopentomutase |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NZ_CP009273 |
4609810 |
4609881 |
72 |
36 [0] |
[0] 36 |
deoB |
phosphopentomutase |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NZ_CP009273 |
= 4609809 | 0 (0.000) | 36 (1.270) |
25/158 |
0.1 |
100% |
coding (390/1224 nt) |
deoB |
phosphopentomutase |
| ? | NZ_CP009273 |
4609882 = |
0 (0.000) | coding (463/1224 nt) |
deoB |
phosphopentomutase |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
CCGTCTGGTCACTGGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATAAACTGGTCGAACGCGCTAATCTGCCGGGTTACCTCGGTAAC > NZ_CP009273/4609714‑4609851
|
CCGTCTGGTCACTGGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAAC < SRR3722099.604774/100‑1 (MQ=60)
CCGTCTGGTCACTGGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGAACACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAAC < SRR3722099.116627/100‑1 (MQ=60)
GTCTGGTCACTGGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTG < SRR3722099.518161/100‑1 (MQ=60)
GGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagca > SRR3722099.809372/1‑90 (MQ=60)
CCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaac < SRR3722099.513248/100‑20 (MQ=60)
GTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaaccggca > SRR3722099.578283/1‑76 (MQ=60)
GTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaaccggca > SRR3722099.165592/1‑76 (MQ=60)
CTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaaccggcaagccgattt > SRR3722099.402477/1‑67 (MQ=60)
CTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaaccggcaagccgattt > SRR3722099.441124/1‑67 (MQ=60)
GTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGGgcgaagagcacatgaaaaccggcaagccgattttc > SRR3722099.775833/1‑65 (MQ=60)
GTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG < SRR3722099.181156/59‑1 (MQ=60)
GGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG < SRR3722099.329404/57‑1 (MQ=60)
GGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG > SRR3722099.107403/1‑55 (MQ=60)
GATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG < SRR3722099.531868/54‑1 (MQ=60)
ATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG > SRR3722099.37452/1‑53 (MQ=60)
ATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG > SRR3722099.535233/1‑53 (MQ=60)
TCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG < SRR3722099.107716/48‑1 (MQ=60)
CGAAAACAGCTTCCCGCAAGAGCTGCTGGATCAACTGG > SRR3722099.787554/1‑38 (MQ=60)
|
CCGTCTGGTCACTGGGAAATTGCCGGTGTCCCGGTTCTGTTTGAGTGGGGATATTTCTCCGATCACGAAAACAGCTTCCCGCAAGAGCTGCTGGATAAACTGGTCGAACGCGCTAATCTGCCGGGTTACCTCGGTAAC > NZ_CP009273/4609714‑4609851
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 26 ≤ ATCG/ATCG < 33 ≤ ATCG/ATCG < 35 ≤ ATCG/ATCG < 41 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |