Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A1 F2 I225 R1
|
227 |
21.4 |
1179702 |
97.1% |
1145490 |
86.5 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NZ_CP009273 |
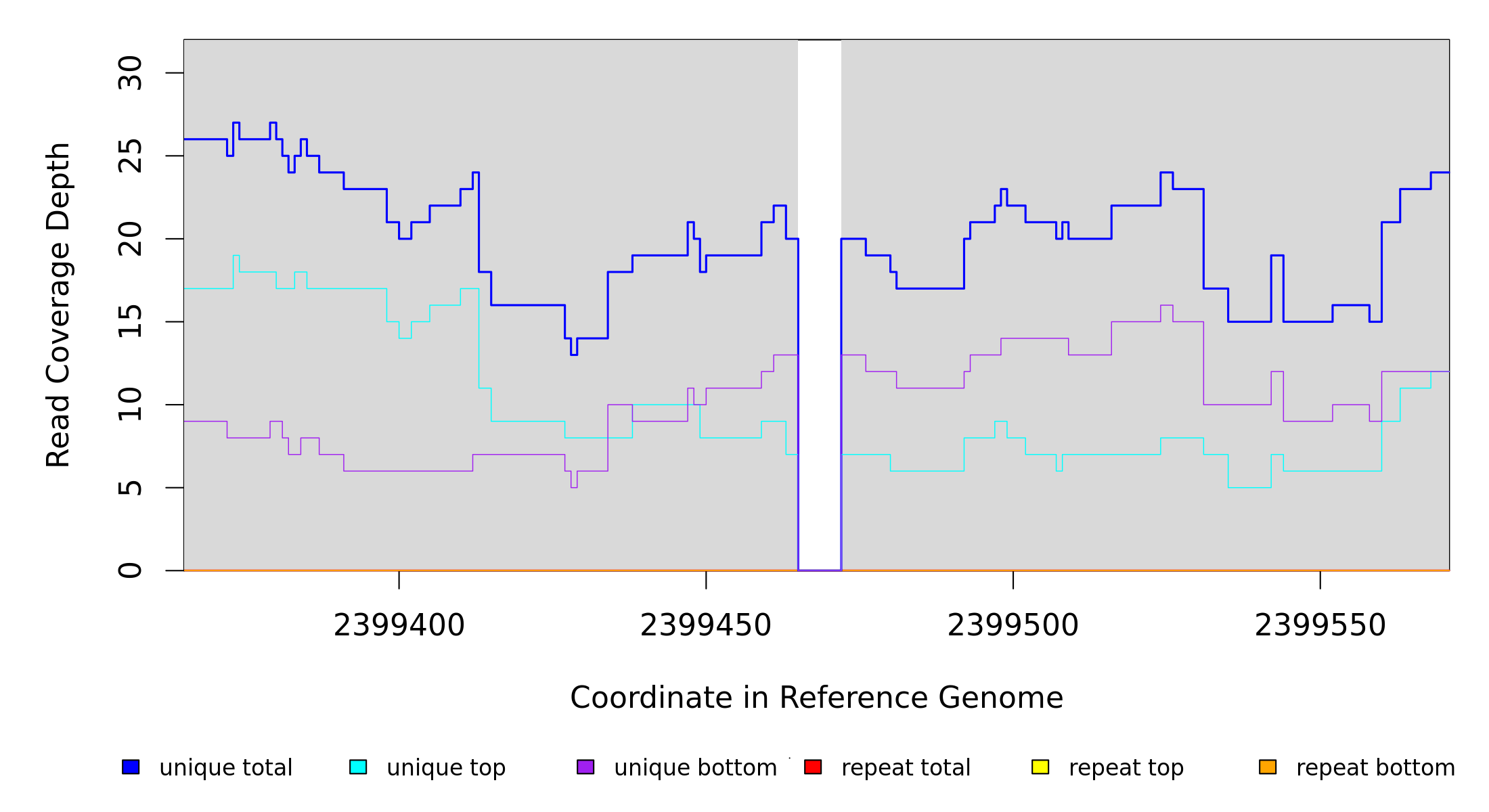
2,399,465 |
Δ7 bp |
coding (650‑656/939 nt) |
lrhA ← |
transcriptional regulator LrhA |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NZ_CP009273 |
2399465 |
2399471 |
7 |
20 [0] |
[0] 20 |
lrhA |
transcriptional regulator LrhA |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NZ_CP009273 |
= 2399464 | 0 (0.000) | 20 (0.950) |
14/172 |
0.3 |
100% |
coding (657/939 nt) |
lrhA |
transcriptional regulator LrhA |
| ? | NZ_CP009273 |
2399472 = |
0 (0.000) | coding (649/939 nt) |
lrhA |
transcriptional regulator LrhA |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATAC > NZ_CP009273/2399384‑2399553
|
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAAT < SRR3722109.583305/100‑1 (MQ=60)
CATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTAT > SRR3722109.140215/1‑100 (MQ=60)
GGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGC > SRR3722109.593223/1‑100 (MQ=60)
GGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGC > SRR3722109.67158/1‑100 (MQ=60)
GCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCT < SRR3722109.114258/100‑1 (MQ=60)
gtgtataagagacagGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722109.137257/85‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722109.360962/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722109.528167/100‑1 (MQ=60)
ACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATAC < SRR3722109.467722/100‑1 (MQ=60)
|
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATAC > NZ_CP009273/2399384‑2399553
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 29 ≤ ATCG/ATCG < 33 ≤ ATCG/ATCG < 35 ≤ ATCG/ATCG < 41 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |