Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A1 F2 I210 R1
|
226 |
19.6 |
1130256 |
95.4% |
1078264 |
84.4 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NZ_CP009273 |
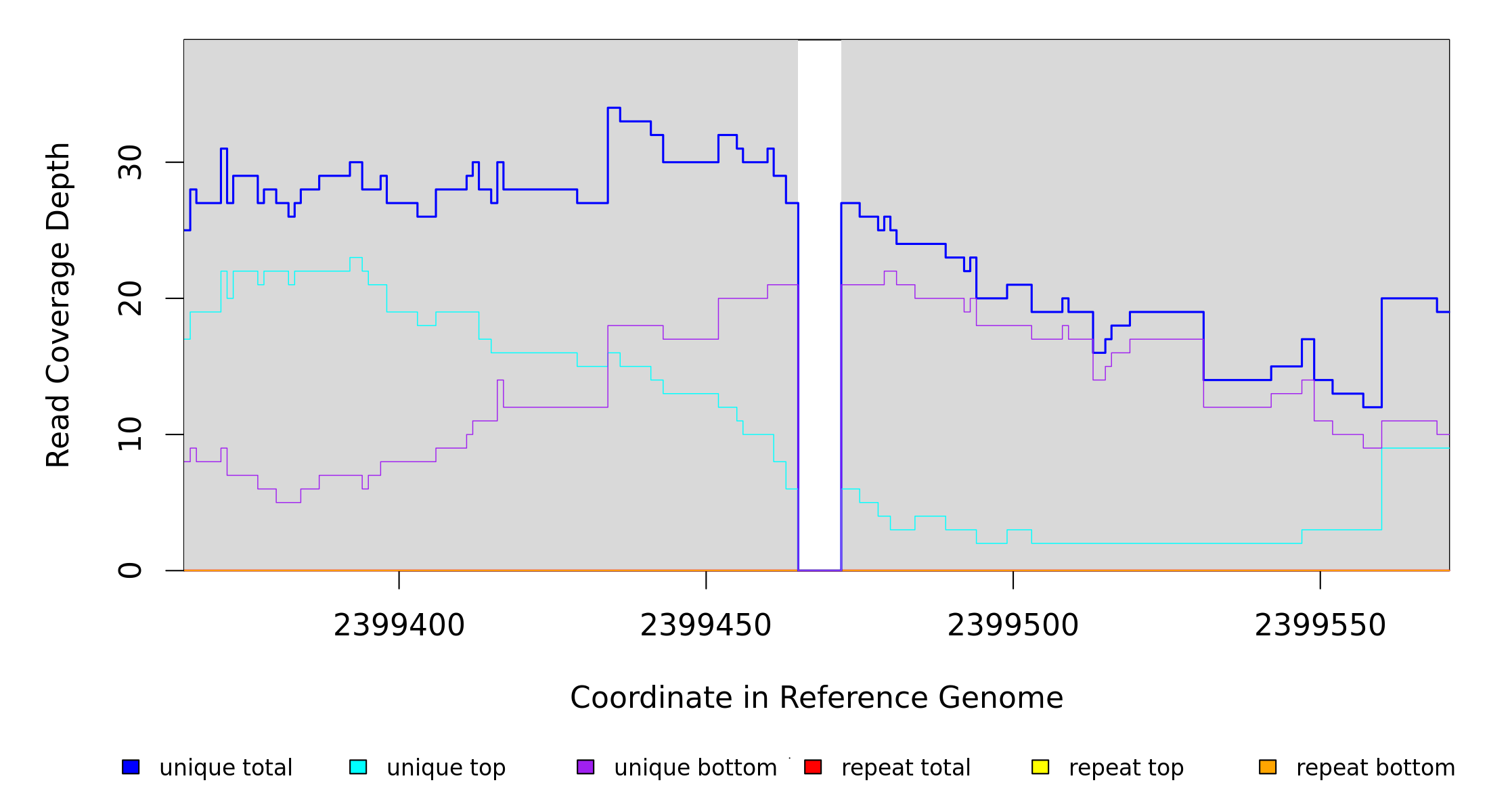
2,399,465 |
Δ7 bp |
coding (650‑656/939 nt) |
lrhA ← |
transcriptional regulator LrhA |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NZ_CP009273 |
2399465 |
2399471 |
7 |
27 [0] |
[0] 27 |
lrhA |
transcriptional regulator LrhA |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NZ_CP009273 |
= 2399464 | 0 (0.000) | 27 (1.400) |
18/166 |
0.1 |
100% |
coding (657/939 nt) |
lrhA |
transcriptional regulator LrhA |
| ? | NZ_CP009273 |
2399472 = |
0 (0.000) | coding (649/939 nt) |
lrhA |
transcriptional regulator LrhA |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACAAGAG > NZ_CP009273/2399384‑2399558
|
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAGGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAAT < SRR3722092.492580/100‑1 (MQ=60)
gcagcgtcagatgtgtataagagacagGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTA < SRR3722092.511239/73‑1 (MQ=60)
GCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTGAGCGTCGCCAGCAC < SRR3722092.499610/100‑1 (MQ=60)
CCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACC < SRR3722092.391008/100‑1 (MQ=60)
CACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATAT < SRR3722092.101129/100‑1 (MQ=60)
CACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATAT < SRR3722092.81432/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722092.126601/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722092.286015/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722092.339656/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722092.49155/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722092.58607/100‑1 (MQ=60)
CGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACAAGAG < SRR3722092.478581/100‑1 (MQ=60)
|
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACAAGAG > NZ_CP009273/2399384‑2399558
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 27 ≤ ATCG/ATCG < 33 ≤ ATCG/ATCG < 35 ≤ ATCG/ATCG < 41 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |