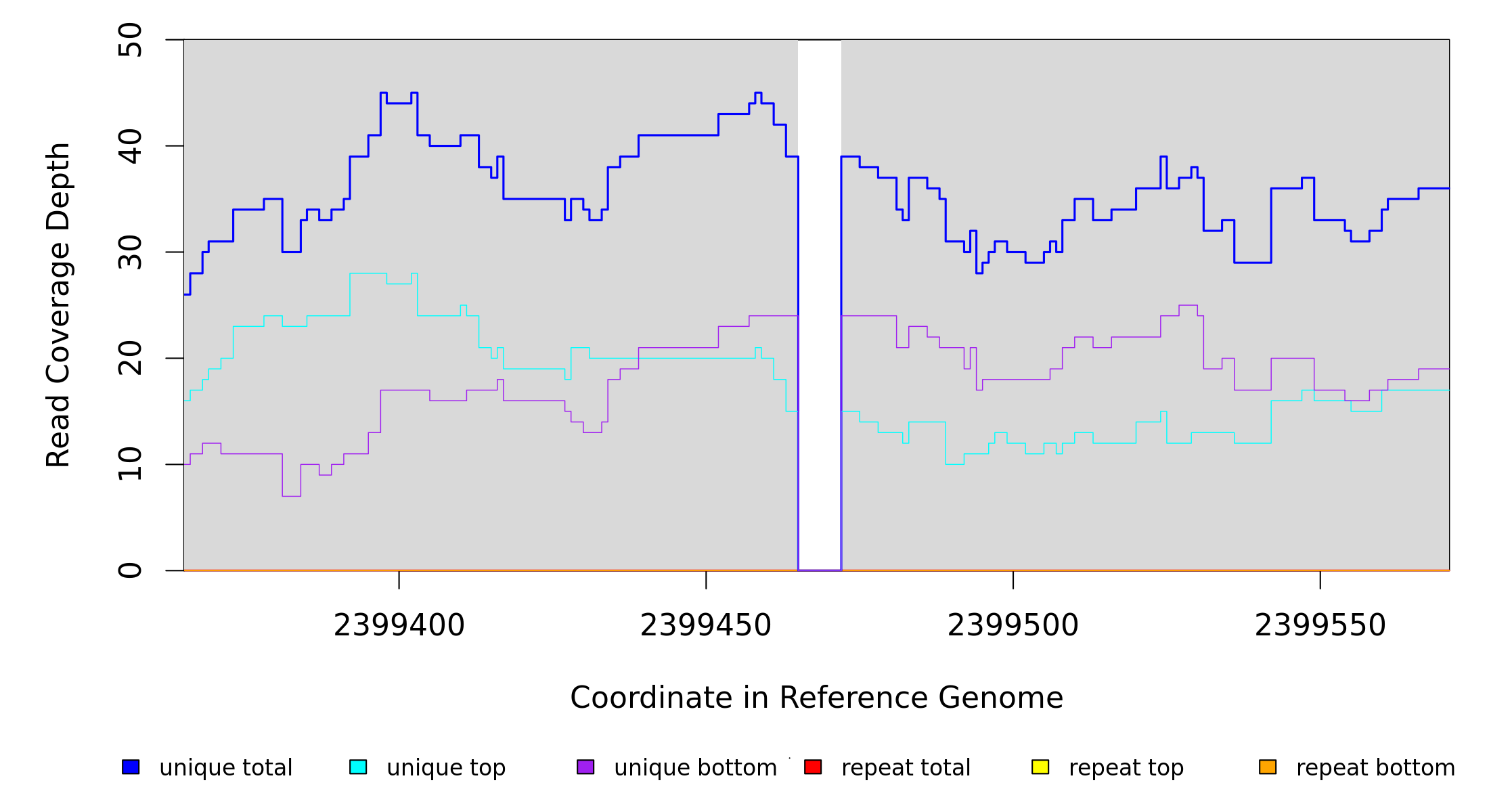
Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A1 F1 I190 R1
|
1 |
32.1 |
1948002 |
94.2% |
1835017 |
83.5 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NZ_CP009273 |
2,399,465 |
Δ7 bp |
coding (650‑656/939 nt) |
lrhA ← |
transcriptional regulator LrhA |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NZ_CP009273 |
2399465 |
2399471 |
7 |
39 [0] |
[0] 39 |
lrhA |
transcriptional regulator LrhA |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NZ_CP009273 |
= 2399464 | 0 (0.000) | 39 (1.200) |
22/164 |
0.3 |
100% |
coding (657/939 nt) |
lrhA |
transcriptional regulator LrhA |
| ? | NZ_CP009273 |
2399472 = |
0 (0.000) | coding (649/939 nt) |
lrhA |
transcriptional regulator LrhA |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
ACACGCAGGTCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACA > NZ_CP009273/2399375‑2399554
|
ACACGCAGGTCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCG > SRR3722066.656519/1‑100 (MQ=60)
GGTCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGA > SRR3722066.653530/1‑100 (MQ=60)
GGTCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGA > SRR3722066.595028/1‑100 (MQ=60)
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAAT < SRR3722066.884446/100‑1 (MQ=60)
TCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAAT < SRR3722066.460890/100‑1 (MQ=60)
TCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCT < SRR3722066.902784/100‑1 (MQ=60)
CATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTT > SRR3722066.387820/1‑100 (MQ=60)
CATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTAT < SRR3722066.38303/100‑1 (MQ=60)
TCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTA < SRR3722066.480103/100‑1 (MQ=60)
TCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTA < SRR3722066.672896/100‑1 (MQ=60)
CAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGC > SRR3722066.544765/1‑100 (MQ=60)
GCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCC > SRR3722066.8360/1‑100 (MQ=60)
GCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCAC < SRR3722066.282837/100‑1 (MQ=60)
CGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCG > SRR3722066.243205/1‑100 (MQ=60)
CGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCG > SRR3722066.172458/1‑100 (MQ=60)
TCACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGA < SRR3722066.312689/100‑1 (MQ=60)
CCCTGCCGCACGAACGGCCGGAAGCGTGGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722066.105344/100‑1 (MQ=60)
CACTGCCGCACGAACGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGAT < SRR3722066.469956/100‑1 (MQ=60)
CGGCCGGAAGCGTCGAG‑‑‑‑‑‑‑AAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACA > SRR3722066.618068/1‑100 (MQ=60)
|
ACACGCAGGTCCGGGCTCATCATCTCAACCGGCCTTGCCGTCACGCCAAGACCGGCTTTCACTGCCGCACGAACGGCCGGAAGCGTCGAGGCGACATAAGCCAGTCGCCATGGAATATCTGCTTTATTAAGCGTCGCCAGCACCATATCGCGAAACGGGCTAGGATCATCCAGCAATACA > NZ_CP009273/2399375‑2399554
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 22 ≤ ATCG/ATCG < 30 ≤ ATCG/ATCG < 34 ≤ ATCG/ATCG < 41 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |