Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A14 F34 I30 R1
|
12 |
87.5 |
2905532 |
96.3% |
2798027 |
145.0 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
CP009273 |
3,360,110 |
Δ10,684 bp |
IS5‑mediated |
[yhcE]–sspA |
[yhcE], yhcF, yhcG, yhcH, nanK, nanE, nanT, nanA, nanR, dcuD, sspB, sspA |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
CP009273 |
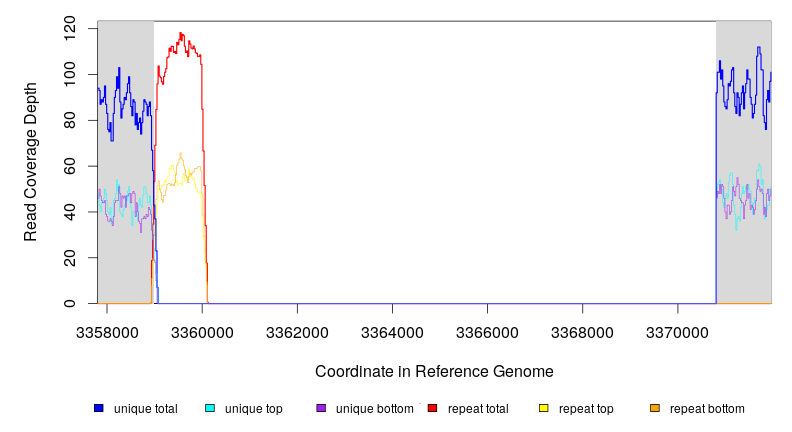
3358981–3360109 |
3370793 |
10685–11813 |
48 [44] |
[0] 90 |
insH1–sspA |
insH1, yhcE, yhcF, yhcG, yhcH, nanK, nanE, nanT, nanA, nanR, dcuD, sspB, sspA |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
CP009273 |
= 3360109 | NA (NA) | 89 (1.020) |
69/288 |
0.2 |
100% |
noncoding (1/1195 nt) |
IS5 |
repeat region |
| ? | CP009273 |
3370794 = |
0 (0.000) | intergenic (‑15/+380) |
sspA/rpsI |
stringent starvation protein A, phage P1 late gene activator, RNAP‑associated acid‑resistance protein, inactive glutathione S‑transferase homolog/30S ribosomal subunit protein S9 |
GATK/CNVnator alignment
N/A