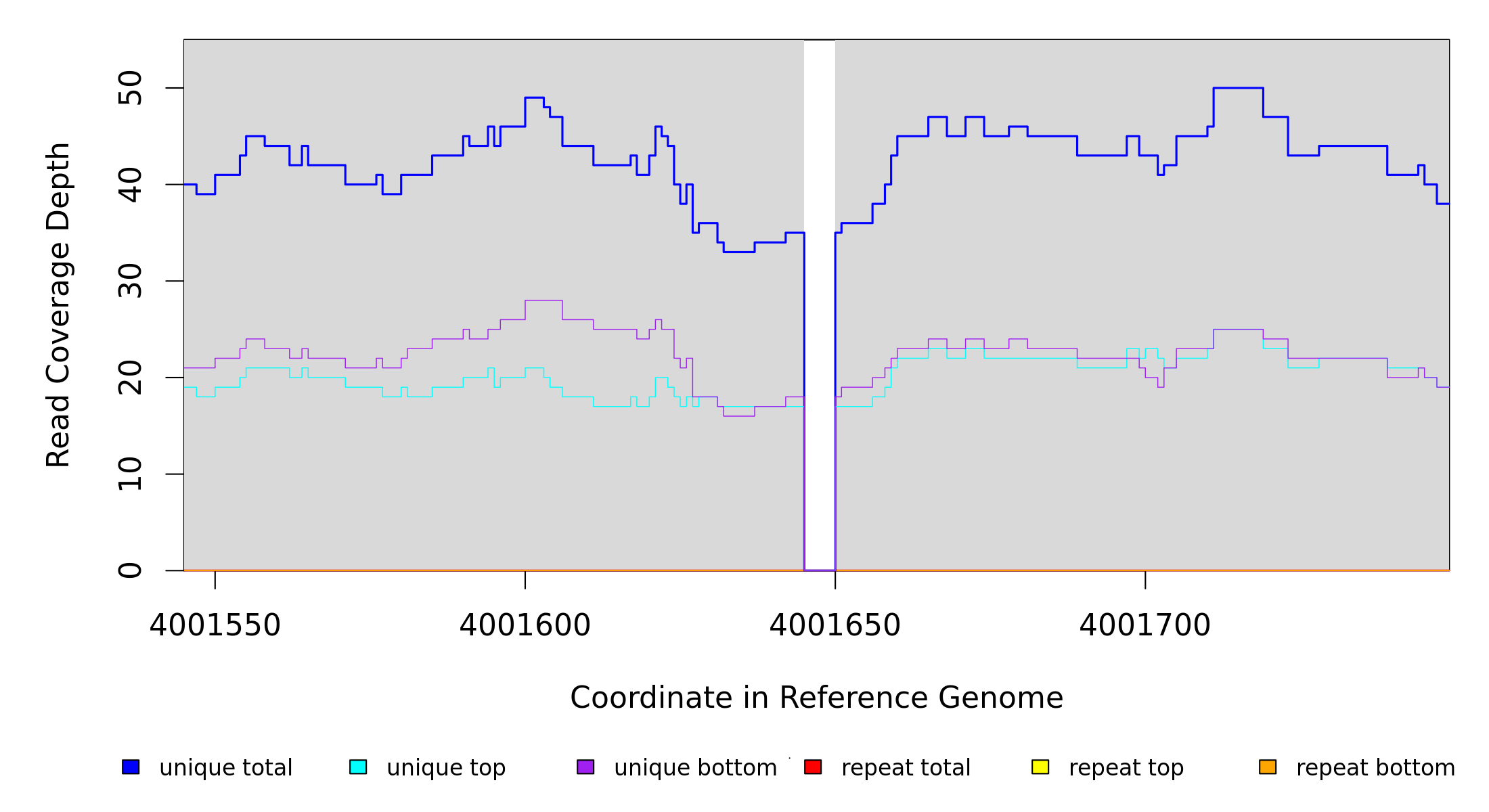
Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A1 F33 I1 R1
|
16 |
30.3 |
1339016 |
95.2% |
1274743 |
112.0 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,001,645 |
Δ5 bp |
coding (220‑224/951 nt) |
corA → |
magnesium/nickel/cobalt transporter |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4001645 |
4001649 |
5 |
35 [0] |
[0] 35 |
corA |
magnesium/nickel/cobalt transporter |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4001644 | 0 (0.000) | 35 (1.160) |
33/222 |
0.1 |
100% |
coding (219/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
| ? | NC_000913 |
4001650 = |
0 (0.000) | coding (225/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
ACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTAT > NC_000913/4001532‑4001775
|
ACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCT > NB501016:228:HFK2FAFXY:1:11208:7918:8281/1‑124 (MQ=60)
ATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCT > NB501016:228:HFK2FAFXY:1:21108:24439:11238/1‑99 (MQ=60)
TGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTG < NB501016:228:HFK2FAFXY:4:11506:23084:4311/132‑1 (MQ=60)
atacgagatacgaattcgtgactggagttcagacgtgtgctcttccgaatctGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGAT < NB501016:228:HFK2FAFXY:2:21211:17567:2928/99‑1 (MQ=60)
GGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTG < NB501016:228:HFK2FAFXY:1:11311:16312:2961/149‑1 (MQ=60)
ACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCacggatggtaaat > NB501016:228:HFK2FAFXY:2:21205:17442:19712/1‑130 (MQ=60)
CCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCG < NB501016:228:HFK2FAFXY:1:11103:11287:6980/90‑1 (MQ=60)
CCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGAT > NB501016:228:HFK2FAFXY:1:11205:15033:2759/1‑131 (MQ=60)
AACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCG < NB501016:228:HFK2FAFXY:1:21311:24660:3508/146‑1 (MQ=60)
GGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCA > NB501016:228:HFK2FAFXY:3:11606:25156:14150/1‑107 (MQ=60)
AAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTG > NB501016:228:HFK2FAFXY:2:11202:17248:19042/1‑67 (MQ=60)
CATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCAgatcggaagaagcacacgtctgaactccagtcacgaattcgtatctcgtatgccgtcttctgcttgaaaaa > NB501016:228:HFK2FAFXY:4:11507:14101:15668/1‑79 (MQ=60)
CATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGG < NB501016:228:HFK2FAFXY:4:11511:9628:9176/99‑1 (MQ=60)
ATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCacggatggtaaatgccacagtgg > NB501016:228:HFK2FAFXY:2:11309:16196:19291/1‑109 (MQ=60)
TCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAA < NB501016:228:HFK2FAFXY:4:21608:14353:13428/45‑1 (MQ=41)
CTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAGCTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTAT > NB501016:228:HFK2FAFXY:1:21207:16244:12368/1‑151 (MQ=60)
CTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGT < NB501016:228:HFK2FAFXY:1:21307:2775:9134/65‑1 (MQ=60)
AAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTC < NB501016:228:HFK2FAFXY:2:11107:2928:4553/94‑1 (MQ=60)
|
ACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTAT > NC_000913/4001532‑4001775
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 21 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |