Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A3 F34 I2 R1
|
15 |
36.8 |
1424632 |
95.4% |
1359098 |
126.1 |
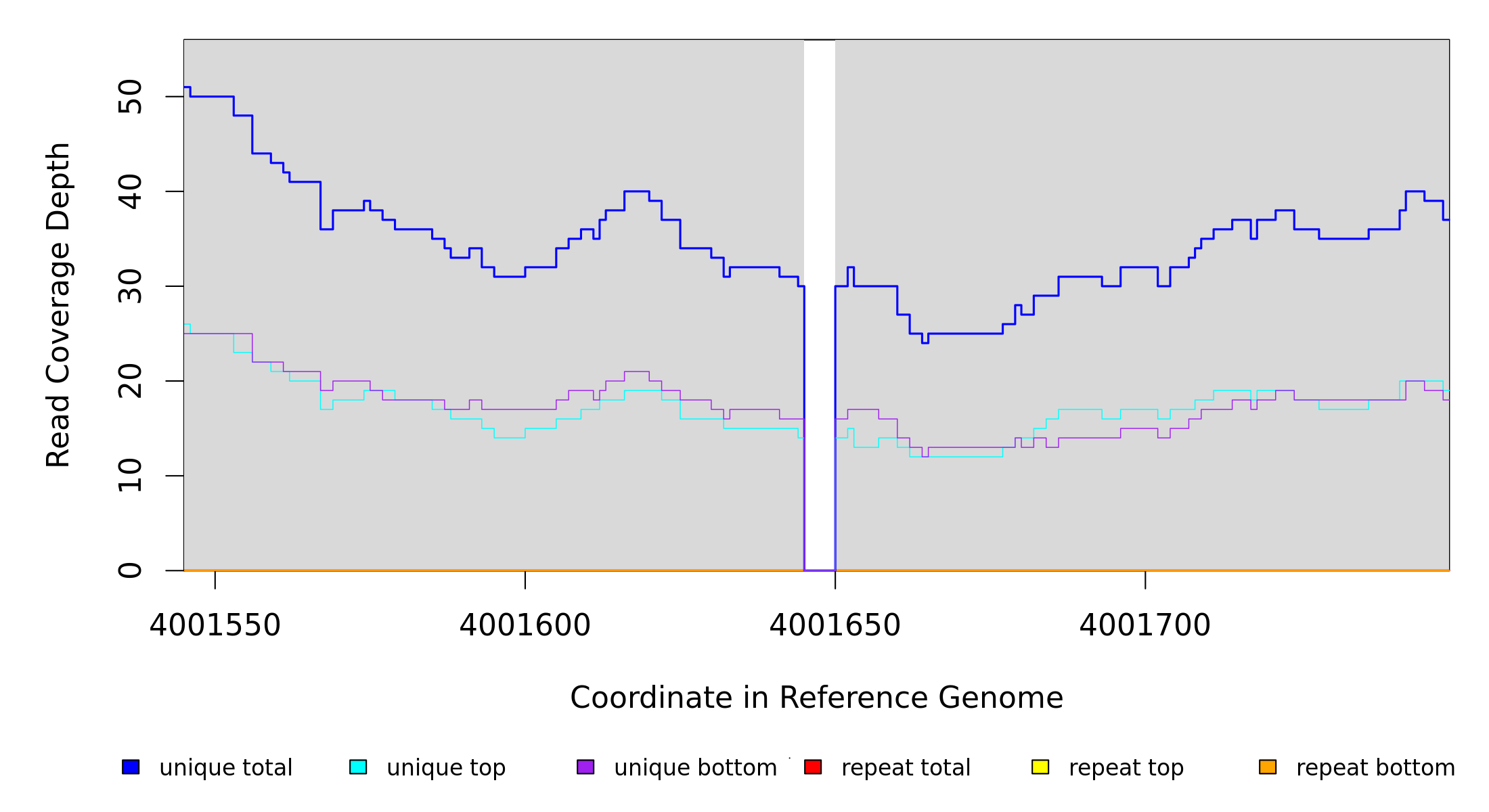
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,001,645 |
Δ5 bp |
coding (220‑224/951 nt) |
corA → |
magnesium/nickel/cobalt transporter |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4001645 |
4001649 |
5 |
30 [0] |
[0] 30 |
corA |
magnesium/nickel/cobalt transporter |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4001644 | 0 (0.000) | 30 (0.820) |
28/250 |
0.4 |
100% |
coding (219/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
| ? | NC_000913 |
4001650 = |
0 (0.000) | coding (225/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
ATGGATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAG > NC_000913/4001506‑4001787
|
ATGGATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCT < NB501016:228:HFK2FAFXY:2:11310:3490:9963/150‑1 (MQ=60)
GATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTT < NB501016:228:HFK2FAFXY:3:21610:21519:17312/151‑1 (MQ=60)
CCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGAT < NB501016:228:HFK2FAFXY:3:21511:4693:15012/151‑1 (MQ=60)
CGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCT > NB501016:228:HFK2FAFXY:2:21303:8071:16461/1‑123 (MQ=60)
AGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACT > NB501016:228:HFK2FAFXY:3:11605:8273:19427/1‑151 (MQ=60)
AGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTC > NB501016:228:HFK2FAFXY:4:11511:1801:12691/1‑120 (MQ=60)
AGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGG > NB501016:228:HFK2FAFXY:4:21606:2623:15157/1‑130 (MQ=60)
CTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTG > NB501016:228:HFK2FAFXY:1:21210:16844:5301/1‑151 (MQ=60)
CATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGGGGCATTTACCATCCGTGATGGTCGGCTGTTTAATATGCGTGAGCGTGAACTGCC > NB501016:228:HFK2FAFXY:4:21401:2519:1141/1‑151 (MQ=60)
AAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCC < NB501016:228:HFK2FAFXY:1:11301:15088:14885/146‑1 (MQ=60)
ATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCG > NB501016:228:HFK2FAFXY:2:21301:19219:3154/1‑151 (MQ=60)
GGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCG < NB501016:228:HFK2FAFXY:2:21307:25701:4338/109‑1 (MQ=60)
GCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTG < NB501016:228:HFK2FAFXY:2:21301:7513:15591/151‑1 (MQ=60)
CGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTG < NB501016:228:HFK2FAFXY:2:21307:22397:8563/98‑1 (MQ=60)
CGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCAGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAG < NB501016:228:HFK2FAFXY:1:21304:14706:14566/151‑1 (MQ=60)
|
ATGGATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAG > NC_000913/4001506‑4001787
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 14 ≤ ATCG/ATCG < 27 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |