Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A4 F30 I1 R1
|
17 |
32.7 |
1267382 |
95.3% |
1207815 |
125.3 |
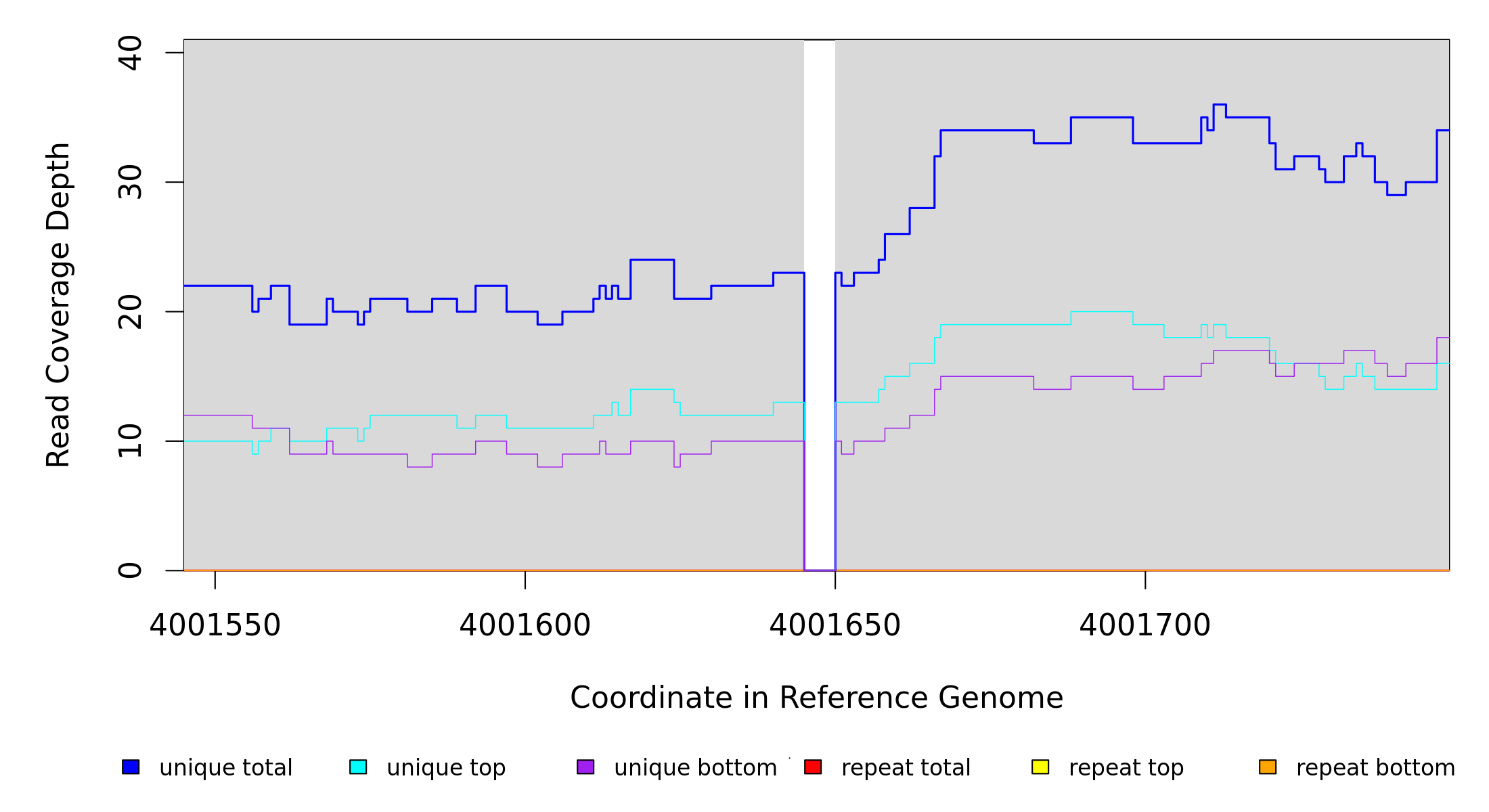
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,001,645 |
Δ5 bp |
coding (220‑224/951 nt) |
corA → |
magnesium/nickel/cobalt transporter |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4001645 |
4001649 |
5 |
23 [0] |
[0] 23 |
corA |
magnesium/nickel/cobalt transporter |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4001644 | 0 (0.000) | 23 (0.710) |
22/248 |
0.6 |
100% |
coding (219/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
| ? | NC_000913 |
4001650 = |
0 (0.000) | coding (225/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
GGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGT > NC_000913/4001527‑4001768
|
GGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCA < NB501016:228:HFK2FAFXY:3:21611:20576:3843/151‑1 (MQ=60)
CGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGC > NB501016:228:HFK2FAFXY:1:21207:5181:16344/1‑151 (MQ=60)
ACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTGTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACAATCCG > NB501016:228:HFK2FAFXY:1:11212:1283:5972/1‑151 (MQ=60)
CAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATG < NB501016:228:HFK2FAFXY:3:21610:15071:2891/149‑1 (MQ=60)
CTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTG > NB501016:228:HFK2FAFXY:2:11302:22872:13136/1‑151 (MQ=60)
CCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTC > NB501016:228:HFK2FAFXY:1:11304:25113:8675/1‑151 (MQ=60)
CCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCG < NB501016:228:HFK2FAFXY:2:11111:12581:2007/151‑1 (MQ=60)
CTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTGCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGG > NB501016:228:HFK2FAFXY:4:11610:4139:9415/1‑126 (MQ=60)
CGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTC > NB501016:228:HFK2FAFXY:3:11407:17979:14763/1‑151 (MQ=60)
GGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTGCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCT < NB501016:228:HFK2FAFXY:4:21404:4995:1888/151‑1 (MQ=60)
CACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGT > NB501016:228:HFK2FAFXY:1:21109:20544:14146/1‑151 (MQ=60)
GTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACT < NB501016:228:HFK2FAFXY:2:11311:19872:19214/78‑1 (MQ=60)
|
GGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGT > NC_000913/4001527‑4001768
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 14 ≤ ATCG/ATCG < 27 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |