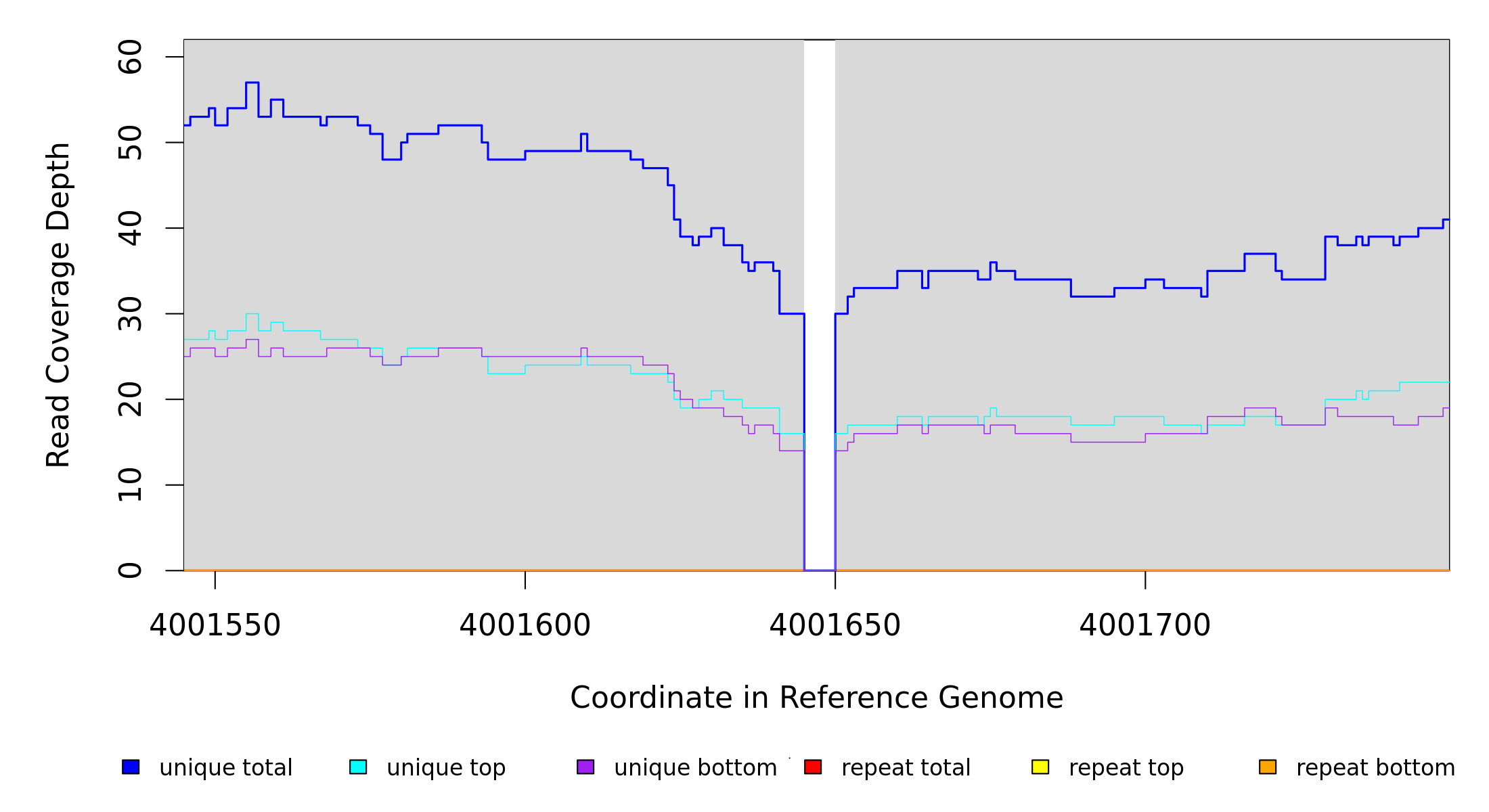
Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A2 F33 I1 R1
|
23 |
35.1 |
1378992 |
95.0% |
1310042 |
124.1 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,001,645 |
Δ5 bp |
coding (220‑224/951 nt) |
corA → |
magnesium/nickel/cobalt transporter |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4001645 |
4001649 |
5 |
30 [0] |
[0] 30 |
corA |
magnesium/nickel/cobalt transporter |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4001644 | 0 (0.000) | 30 (0.870) |
28/246 |
0.4 |
100% |
coding (219/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
| ? | NC_000913 |
4001650 = |
0 (0.000) | coding (225/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
GATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCG > NC_000913/4001509‑4001784
|
GATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATCT < NB501016:228:HFK2FAFXY:4:11601:5526:1455/138‑1 (MQ=60)
GTCGAACCGGACTACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGACTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCG < NB501016:228:HFK2FAFXY:2:11107:20809:19924/151‑1 (MQ=60)
cccccccccccccgtttttttttcacgcagaagacggcatacgcgctgcgcattagagacgggagtgcagacgtgggttcttccgatctGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAG < NB501016:228:HFK2FAFXY:4:11511:12438:8490/62‑1 (MQ=60)
ACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGA < NB501016:228:HFK2FAFXY:1:21201:10271:7577/151‑1 (MQ=60)
GACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAAC < NB501016:228:HFK2FAFXY:4:11504:10927:7028/147‑1 (MQ=60)
GCGCGTACAATCTGATCTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACTACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTG < NB501016:228:HFK2FAFXY:3:21509:24631:16539/117‑1 (MQ=60)
ACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGC > NB501016:228:HFK2FAFXY:2:11103:6129:2674/1‑148 (MQ=60)
ATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTT < NB501016:228:HFK2FAFXY:3:11508:2883:9862/106‑1 (MQ=60)
CCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCC < NB501016:228:HFK2FAFXY:1:11310:12619:5870/116‑1 (MQ=60)
CAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGT < NB501016:228:HFK2FAFXY:1:21310:23837:10760/151‑1 (MQ=60)
GCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTT < NB501016:228:HFK2FAFXY:1:21307:2475:8167/151‑1 (MQ=60)
ACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTGTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGCTCACGCCGG < NB501016:228:HFK2FAFXY:4:21507:14060:16008/105‑1 (MQ=60)
CCCGCCCGGAACTGGAAGACATCGAAGCCTCGGCACGTTTCTTTGAAGACGACGCCGGCCTGCAT‑‑‑‑‑CTCCTACTTCTTCTTGGAAGATGCGGAAGATCACGCCGGTCACTCCACGGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTC > NB501016:228:HFK2FAFXY:4:21510:26588:6082/1‑151 (MQ=60)
ATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGG > NB501016:228:HFK2FAFXY:2:21210:17391:15361/1‑109 (MQ=60)
GACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCC > NB501016:228:HFK2FAFXY:3:11611:10270:8319/1‑151 (MQ=60)
CGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCG > NB501016:228:HFK2FAFXY:2:21203:10926:15294/1‑151 (MQ=60)
|
GATTGATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCG > NC_000913/4001509‑4001784
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 14 ≤ ATCG/ATCG < 21 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |