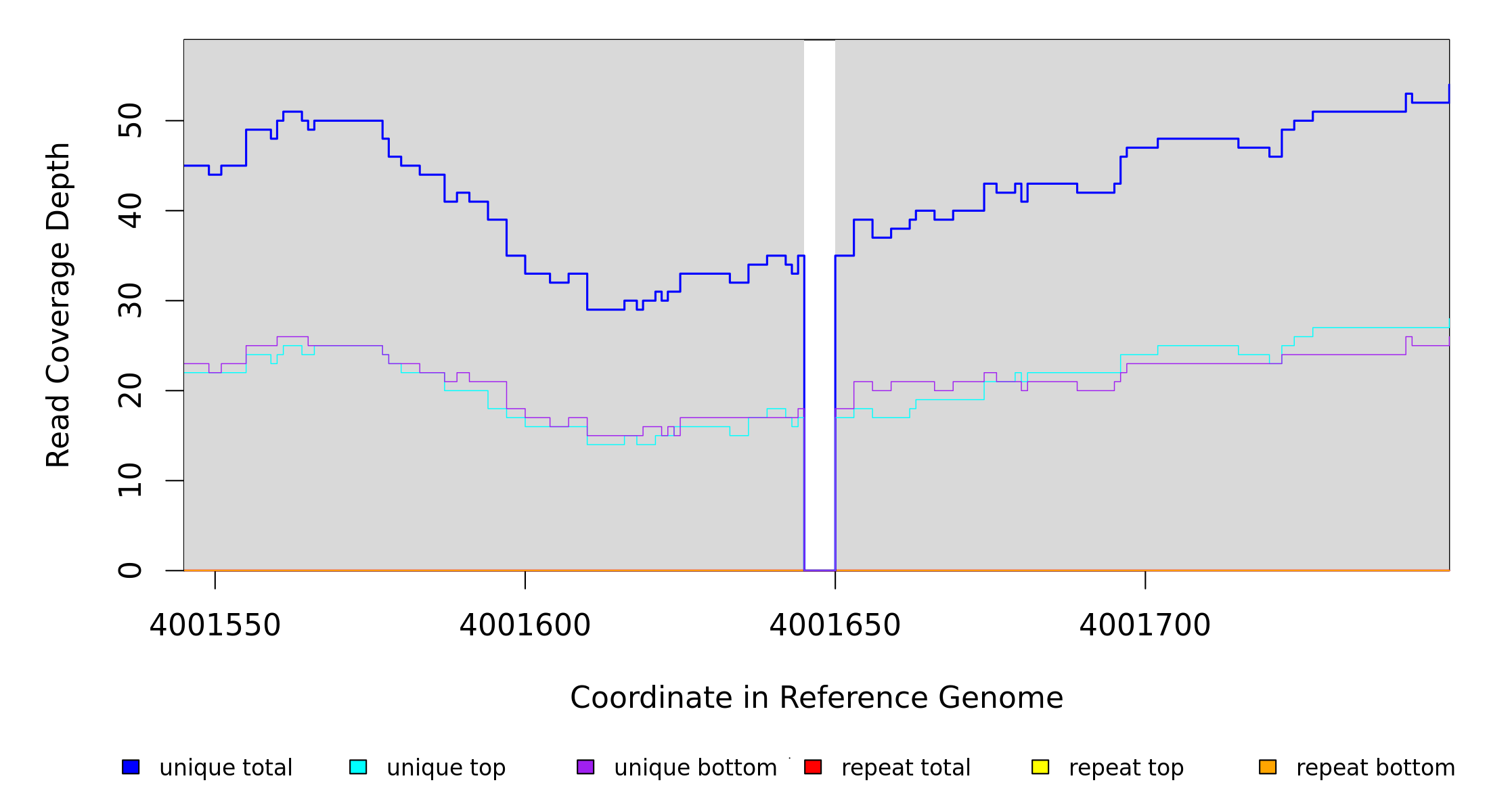
Sample Resequencing Stats
Note: The mutation counts shown below represent unfiltered mutation sets.
| ALE, Flask, Isolate |
Predicted Mutations |
Mean Coverage |
Total Reads |
Percent Mapped |
Mapped Reads |
Average Read Length |
|
A4 F9 I1 R1
|
15 |
36.8 |
1385066 |
95.5% |
1322738 |
129.6 |
Breseq alignment
BRESEQ :: Evidence
|
| evidence |
seq id |
position |
mutation |
annotation |
gene |
description |
| MC JC |
NC_000913 |
4,001,645 |
Δ5 bp |
coding (220‑224/951 nt) |
corA → |
magnesium/nickel/cobalt transporter |
|
| | | | seq id |
start |
end |
size |
←reads |
reads→ |
gene |
description |
|---|
| * |
* |
÷ |
NC_000913 |
4001645 |
4001649 |
5 |
35 [0] |
[0] 35 |
corA |
magnesium/nickel/cobalt transporter |
|
| |
seq id |
position |
reads (cov) |
reads (cov) |
score |
skew |
freq |
annotation |
gene |
product |
| * |
? |
NC_000913 |
= 4001644 | 0 (0.000) | 35 (0.960) |
33/258 |
0.2 |
100% |
coding (219/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
| ? | NC_000913 |
4001650 = |
0 (0.000) | coding (225/951 nt) |
corA |
magnesium/nickel/cobalt transporter |
GATK/CNVnator alignment
BRESEQ :: bam2aln output
GATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAGCC > NC_000913/4001513‑4001789
|
GATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGA < NB501016:228:HFK2FAFXY:3:11404:15507:17666/150‑1 (MQ=60)
CGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGA < NB501016:228:HFK2FAFXY:2:21211:15840:5987/151‑1 (MQ=60)
GCGGACAATCTGACCTTGGCCAGAGCCTGGCAACCCGCCCGGAACAGGAAGACAGCGAAGCATCGGCACGGTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGG < NB501016:228:HFK2FAFXY:4:21604:24817:16078/151‑1 (MQ=60)
ATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGCCATCGAAGCATCGGCACGTTTCTGTGAAGCCGACGACGGCCGGCA‑‑‑‑‑GCTCCTTCGTCTTCTGTGCAGATGCGGAAGAT > NB501016:228:HFK2FAFXY:4:11504:1219:2251/1‑122 (MQ=60)
AACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCC < NB501016:228:HFK2FAFXY:3:21608:9018:1379/123‑1 (MQ=60)
ACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCG > NB501016:228:HFK2FAFXY:3:21505:19900:13390/1‑151 (MQ=60)
GCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGCCGGCCTGCAT‑‑‑‑‑CGCCTTCTTCTTCGTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATG > NB501016:228:HFK2FAFXY:4:11508:22738:18870/1‑151 (MQ=60)
GAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAG < NB501016:228:HFK2FAFXY:2:11210:24542:12011/151‑1 (MQ=60)
CGTTTCTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTAT > NB501016:228:HFK2FAFXY:3:11609:12694:14506/1‑151 (MQ=60)
CTTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTAT > NB501016:228:HFK2FAFXY:3:11405:8248:19262/1‑151 (MQ=60)
TTTGAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAAC < NB501016:228:HFK2FAFXY:2:21307:1553:15051/125‑1 (MQ=60)
GAAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTT < NB501016:228:HFK2FAFXY:1:11303:13009:20211/132‑1 (MQ=60)
AAGACGACGACGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCacgctcacg < NB501016:228:HFK2FAFXY:3:21612:14811:15447/142‑10 (MQ=60)
CGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAGCC > NB501016:228:HFK2FAFXY:1:11106:26598:19428/1‑150 (MQ=60)
CGGCCTGCAT‑‑‑‑‑CTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAGCC > NB501016:228:HFK2FAFXY:1:21107:14941:7754/1‑150 (MQ=60)
|
GATCTTGTCGAACCGGACGACGACGAGCGACTGCGCGTACAATCTGAACTTGGCCAGAGCCTGGCAACCCGCCCGGAACTGGAAGACATCGAAGCATCGGCACGTTTCTTTGAAGACGACGACGGCCTGCATATTCACTCCTTCTTCTTCTTTGAAGATGCGGAAGATCACGCCGGTAACTCCACTGTGGCATTTACCATCCGTGATGGTCGTCTGTTTACTCTGCGTGAGCGTGAACTGCCCGCTTTTCGTCTGTATCGTATGCGTGCCCGTAGCC > NC_000913/4001513‑4001789
|
| Alignment Legend |
|---|
Aligned base mismatch/match (shaded by quality score): ATCG/ATCG < 0 ≤ ATCG/ATCG < 14 ≤ ATCG/ATCG < 27 ≤ ATCG/ATCG < 32 ≤ ATCG/ATCG < 36 ≤ ATCG/ATCG |
Unaligned base: atcg Masked matching base: atcg Alignment gap: ‑ Deleted base: ‑ |